Epidemiology of Non-SARS-CoV2 Human Coronaviruses (HCoVs) in People Presenting with Influenza-like Illness (ILI) or Severe Acute Respiratory Infections (SARI) in Senegal from 2012 to 2020
- PMID: 36680061
- PMCID: PMC9864203
- DOI: 10.3390/v15010020
Epidemiology of Non-SARS-CoV2 Human Coronaviruses (HCoVs) in People Presenting with Influenza-like Illness (ILI) or Severe Acute Respiratory Infections (SARI) in Senegal from 2012 to 2020
Abstract
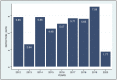
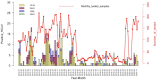
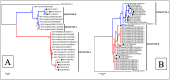
In addition to emerging coronaviruses (SARS-CoV, MERS, SARS-CoV-2), there are seasonal human coronaviruses (HCoVs): HCoV-OC43, HCoV-229E, HCoV-NL63 and HCoV-HKU1. With a wide distribution around the world, HCoVs are usually associated with mild respiratory disease. In the elderly, young children and immunocompromised patients, more severe or even fatal respiratory infections may be observed. In Africa, data on seasonal HCoV are scarce. This retrospective study investigated the epidemiology and genetic diversity of seasonal HCoVs during nine consecutive years of influenza-like illness surveillance in Senegal. Nasopharyngeal swabs were collected from ILI outpatients or from SARI hospitalized patients. HCoVs were diagnosed by qRT-PCR and the positive samples were selected for molecular characterization. Among 9337 samples tested for HCoV, 406 (4.3%) were positive: 235 (57.9%) OC43, 102 (25.1%) NL63, 58 (14.3%) 229E and 17 (4.2%) HKU1. The four types circulated during the study period and a peak was noted between November and January. Children under five were the most affected. Co-infections were observed between HCoV types (1.2%) or with other viruses (76.1%). Genetically, HCoVs types showed diversity. The results highlighted that the impact of HCoVs must be taken into account in public health; monitoring them is therefore particularly necessary both in the most sensitive populations and in animals.
Keywords: HCoV-229E; HCoV-HKU1; HCoV-NL63; HCoV-OC43; ILI; SARI; coronavirus characterization; epidemiology; human coronaviruses; respiratory infections.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Monitoring of human coronaviruses in Belgian primary care and hospitals, 2015-20: a surveillance study.Lancet Microbe. 2021 Mar;2(3):e105-e114. doi: 10.1016/S2666-5247(20)30221-4. Epub 2021 Jan 27. Lancet Microbe. 2021. PMID: 33937883 Free PMC article.
-
Continuous evolution and emerging lineage of seasonal human coronaviruses: A multicenter surveillance study.J Med Virol. 2023 Jun;95(6):e28861. doi: 10.1002/jmv.28861. J Med Virol. 2023. PMID: 37310144
-
An overview on the seven pathogenic human coronaviruses.Rev Med Virol. 2022 Mar;32(2):e2282. doi: 10.1002/rmv.2282. Epub 2021 Aug 2. Rev Med Virol. 2022. PMID: 34339073 Review.
-
Epidemiological and co-infection characteristics of common human coronaviruses in Shanghai, 2015-2020: a retrospective observational study.Emerg Microbes Infect. 2021 Dec;10(1):1660-1668. doi: 10.1080/22221751.2021.1965498. Emerg Microbes Infect. 2021. PMID: 34350810 Free PMC article.
-
Seasonality of Common Human Coronaviruses, United States, 2014-20211.Emerg Infect Dis. 2022 Oct;28(10):1970-1976. doi: 10.3201/eid2810.220396. Epub 2022 Aug 25. Emerg Infect Dis. 2022. PMID: 36007923 Free PMC article. Review.
Cited by
-
Prevalence and clinical impact of mono- and co-infections with endemic coronaviruses 229E, OC43, NL63, and HKU-1 during the COVID-19 pandemic.Heliyon. 2024 Apr 7;10(7):e29258. doi: 10.1016/j.heliyon.2024.e29258. eCollection 2024 Apr 15. Heliyon. 2024. PMID: 38623185 Free PMC article.
-
Epidemiology of Human Seasonal Coronaviruses Among People With Mild and Severe Acute Respiratory Illness in Blantyre, Malawi, 2011-2017.J Infect Dis. 2024 Aug 16;230(2):e363-e373. doi: 10.1093/infdis/jiad587. J Infect Dis. 2024. PMID: 38365443 Free PMC article.
-
Enhancing epidemic preparedness: a data-driven system for managing respiratory infections.BMC Infect Dis. 2025 Feb 3;25(1):159. doi: 10.1186/s12879-025-10528-y. BMC Infect Dis. 2025. PMID: 39901070 Free PMC article.
-
Epidemiologic and clinical updates on viral infections in Saudi Arabia.Saudi Pharm J. 2024 Jul;32(7):102126. doi: 10.1016/j.jsps.2024.102126. Epub 2024 Jun 8. Saudi Pharm J. 2024. PMID: 38966679 Free PMC article.
-
Whole Genome Sequencing and Genetic Diversity of Respiratory Viruses Detected in Children With Acute Respiratory Infections: A One-Year Cross-Sectional Study in Senegal.J Med Virol. 2025 Apr;97(4):e70342. doi: 10.1002/jmv.70342. J Med Virol. 2025. PMID: 40207925 Free PMC article.
References
-
- Kin N., Miszczak F., Diancourt L., Caro V., Moutou F., Vabret A., Gouilh M.A. Comparative molecular epidemiology of two closely related coronaviruses, bovine coronavirus (BCoV) and human coronavirus OC43 (HCoV-OC43), reveals a different evolutionary pattern. Infect. Genet. Evol. 2016;40:186–191. doi: 10.1016/j.meegid.2016.03.006. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

