Determinants of Retroviral Integration and Implications for Gene Therapeutic MLV-Based Vectors and for a Cure for HIV-1 Infection
- PMID: 36680071
- PMCID: PMC9861059
- DOI: 10.3390/v15010032
Determinants of Retroviral Integration and Implications for Gene Therapeutic MLV-Based Vectors and for a Cure for HIV-1 Infection
Abstract
To complete their replication cycle, retroviruses need to integrate a DNA copy of their RNA genome into a host chromosome. Integration site selection is not random and is driven by multiple viral and cellular host factors specific to different classes of retroviruses. Today, overwhelming evidence from cell culture, animal experiments and clinical data suggests that integration sites are important for retroviral replication, oncogenesis and/or latency. In this review, we will summarize the increasing knowledge of the mechanisms underlying the integration site selection of the gammaretrovirus MLV and the lentivirus HIV-1. We will discuss how host factors of the integration site selection of retroviruses may steer the development of safer viral vectors for gene therapy. Next, we will discuss how altering the integration site preference of HIV-1 using small molecules could lead to a cure for HIV-1 infection.
Keywords: HIV-1; MLV; host factors; integration site selection; latency; retroviral integration; viral vector.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
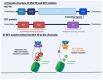
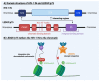

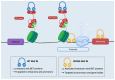
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

