Inactivation of Ebola Virus and SARS-CoV-2 in Cell Culture Supernatants and Cell Pellets by Gamma Irradiation
- PMID: 36680083
- PMCID: PMC9866162
- DOI: 10.3390/v15010043
Inactivation of Ebola Virus and SARS-CoV-2 in Cell Culture Supernatants and Cell Pellets by Gamma Irradiation
Abstract
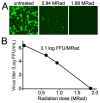
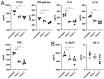
Viral pathogens with the potential to cause widespread disruption to human health and society continue to emerge or re-emerge around the world. Research on such viruses often involves high biocontainment laboratories (BSL3 or BSL4), but the development of diagnostics, vaccines and therapeutics often uses assays that are best performed at lower biocontainment. Reliable inactivation is necessary to allow removal of materials to these spaces and to ensure personnel safety. Here, we validate the use of gamma irradiation to inactivate culture supernatants and pellets of cells infected with a representative member of the Filovirus and Coronavirus families. We show that supernatants and cell pellets containing SARS-CoV-2 are readily inactivated with 1.9 MRad, while Ebola virus requires higher doses of 2.6 MRad for supernatants and 3.8 MRad for pellets. While these doses of radiation inactivate viruses, proinflammatory cytokines that are common markers of virus infection are still detected with low losses. The doses required for virus inactivation of supernatants are in line with previously reported values, but the inactivation of cell pellets has not been previously reported and enables new approaches for analysis of protein-based host responses to infection.
Keywords: Ebola virus; SARS-CoV-2; coronavirus; filovirus; inactivation; radiation treatment; sterilization.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures

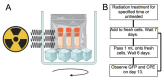
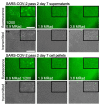

References
-
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., et al. Genomic Characterisation and Epidemiology of 2019 Novel Coronavirus: Implications for Virus Origins and Receptor Binding. Lancet Lond. Engl. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

