Molecular Epidemiology of HIV-1 in Ghana: Subtype Distribution, Drug Resistance and Coreceptor Usage
- PMID: 36680168
- PMCID: PMC9865111
- DOI: 10.3390/v15010128
Molecular Epidemiology of HIV-1 in Ghana: Subtype Distribution, Drug Resistance and Coreceptor Usage
Abstract
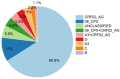
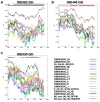
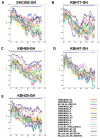
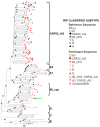
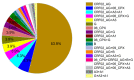
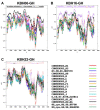
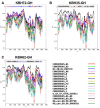
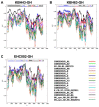
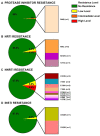
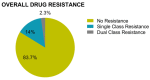
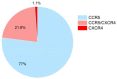
The greatest HIV-1 genetic diversity is found in West/Central Africa due to the pandemic’s origins in this region, but this diversity remains understudied. We characterized HIV-1 subtype diversity (from both sub-genomic and full-genome viral sequences), drug resistance and coreceptor usage in 103 predominantly (90%) antiretroviral-naive individuals living with HIV-1 in Ghana. Full-genome HIV-1 subtyping confirmed the circulating recombinant form CRF02_AG as the dominant (53.9%) subtype in the region, with the complex recombinant 06_cpx (4%) present as well. Unique recombinants, most of which were mosaics containing CRF02_AG and/or 06_cpx, made up 37% of sequences, while “pure” subtypes were rare (<6%). Pretreatment resistance to at least one drug class was observed in 17% of the cohort, with NNRTI resistance being the most common (12%) and INSTI resistance being relatively rare (2%). CXCR4-using HIV-1 sequences were identified in 23% of participants. Overall, our findings advance our understanding of HIV-1 molecular epidemiology in Ghana. Extensive HIV-1 genetic diversity in the region appears to be fueling the ongoing creation of novel recombinants, the majority CRF02_AG-containing, in the region. The relatively high prevalence of pretreatment NNRTI resistance but low prevalence of INSTI resistance supports the use of INSTI-based first-line regimens in Ghana.
Keywords: Ghana; HIV; HIV-1; coreceptor usage; molecular epidemiology; pretreatment drug resistance; subtype diversity.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the result.
Figures
References
-
- Nweze J., Eke I., Nweze E. HIV/AIDS in sub-Saharan Africa: Current status, challenges and prospects. Asian Pac. J. Trop. Dis. 2017;7:239–256. doi: 10.12980/apjtd.7.2017D6-366. - DOI
-
- UNAIDS Global HIV & AIDS Statistics—Fact Sheet. [(accessed on 1 September 2022)]. Available online: https://www.unaids.org/en/resources/fact-sheet.
-
- Hemelaar J., Elangovan R., Yun J., Dickson-Tetteh L., Fleminger I., Kirtley S., Williams B., Gouws-Williams E., Ghys P.D., Abimiku A.l.G., et al. Global and regional molecular epidemiology of HIV-1, 1990–2015: A systematic review, global survey, and trend analysis. Lancet Infect. Dis. 2019;19:143–155. doi: 10.1016/S1473-3099(18)30647-9. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

