Phage Adsorption to Gram-Positive Bacteria
- PMID: 36680236
- PMCID: PMC9863714
- DOI: 10.3390/v15010196
Phage Adsorption to Gram-Positive Bacteria
Abstract
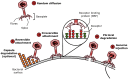
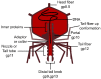
The phage life cycle is a multi-stage process initiated by the recognition and attachment of the virus to its bacterial host. This adsorption step depends on the specific interaction between bacterial structures acting as receptors and viral proteins called Receptor Binding Proteins (RBP). The adsorption process is essential as it is the first determinant of phage host range and a sine qua non condition for the subsequent conduct of the life cycle. In phages belonging to the Caudoviricetes class, the capsid is attached to a tail, which is the central player in the adsorption as it comprises the RBP and accessory proteins facilitating phage binding and cell wall penetration prior to genome injection. The nature of the viral proteins involved in host adhesion not only depends on the phage morphology (i.e., myovirus, siphovirus, or podovirus) but also the targeted host. Here, we give an overview of the adsorption process and compile the available information on the type of receptors that can be recognized and the viral proteins taking part in the process, with the primary focus on phages infecting Gram-positive bacteria.
Keywords: Gram-positive host; adsorption; phage; receptor; receptor binding protein; tail.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







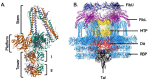
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

