NFYAv1 is a Tumor-Promoting Transcript Associated with Poor Prognosis of Hepatocellular Carcinoma
- PMID: 36680333
- PMCID: PMC9875548
- DOI: 10.12659/MSM.938410
NFYAv1 is a Tumor-Promoting Transcript Associated with Poor Prognosis of Hepatocellular Carcinoma
Abstract
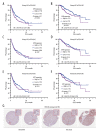
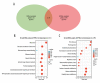
BACKGROUND Nuclear Transcription Factor Y Subunit Alpha (NFYA), together with NFYB and NFYC, form a sequence-specific heterotrimeric nuclear transcription factor (NFY), but their functional role in hepatocellular carcinoma (HCC) is still unclear. In this study, we explored the association between the NFY subunit genes and the survival of primary hepatocellular carcinoma (HCC) patients in The Cancer Genome Atlas (TCGA). The transcript-specific effect on HCC cell growth was studied. MATERIAL AND METHODS RNA-seq data from the Genotype-Tissue Expression Project (GTEx) and TCGA were analyzed in combination. In vitro cellular and molecular studies were conducted using SK-Hep-1 and Hep3B cells. Pearson's correlation coefficients were calculated to assess correlations. Welch's unpaired t test and one-way ANOVA with post hoc Tukey's multiple comparisons were performed. Kaplan-Meier (K-M) survival curves were assessed by conducting log-rank (Mantel-Cox) test. RESULTS NFYA was the only prognosis-related gene. Among the 2 splicing transcripts of NFYA, the long isoform (NFYAv1, NM_002505.5) but not the short-form (NFYAv2, NM_021705.4) was significantly associated with worse progression-free survival (PFS) (high [n=179] vs low [n=179], HR: 1.657, 95% CI: 1.228-2.235, P<0.001) and disease-specific survival (DSS) (high [n=175] vs low [n=175], HR: 1.986, 95% CI: 1.269-3.108, P<0.001) in HCC patients. GO/KEGG analysis in TCGA confirmed that NFYAv1 and NFYAv2 co-expressed (|Pearson's r|≥0.6) genes in primary HCC patients were enriched in quite different GO/KEGG terms. NFYAv1 knockdown significantly decreased cell viability and increased G0/G1 cell cycle arrest. The shRNA only targeting NFYAv1 had a significantly stronger growth-inhibiting effect than the shRNA targeting both NFYAv1 and NFYAv2. CONCLUSIONS This study showed that NFYAv1 is a tumor-promoting transcript associated with poor prognosis of HCC.
Conflict of interest statement
Figures


References
-
- Gurtner A, Manni I, Piaggio G. NF-Y in cancer: Impact on cell transformation of a gene essential for proliferation. Biochim Biophys Acta Gene Regul Mech. 2017;1860(5):604–16. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

