Uncovering the determinants of model Escherichia coli strain C600 susceptibility and resistance to lytic T4-like and T7-like phage
- PMID: 36681192
- PMCID: PMC10194157
- DOI: 10.1016/j.virusres.2023.199048
Uncovering the determinants of model Escherichia coli strain C600 susceptibility and resistance to lytic T4-like and T7-like phage
Abstract
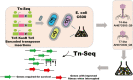
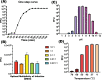
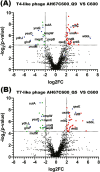
As antimicrobial resistance (AMR) continues to increase, the therapeutic use of phages has re-emerged as an attractive alternative. However, knowledge of phage resistance development and bacterium-phage interaction complexity are still not fully interpreted. In this study, two lytic T4-like and T7-like phage infecting model Escherichia coli strain C600 are selected, and host genetic determinants involved in phage susceptibility and resistance are also identified using TraDIS strategy. Isolation and identification of the lytic T7-like show that though it belongs to the phage T7 family, genes encoding replication and transcription protein exhibit high differences. The TraDIS results identify a huge number of previously unidentified genes involved in phage infection, and a subset (six in susceptibility and nine in resistance) are shared under pressure of the two kinds of lytic phage. Susceptible gene wbbL has the highest value and implies the important role in phage susceptibility. Importantly, two susceptible genes QseE (QseE/QseF) and RstB (RstB/RstA), encoding the similar two-component system sensor histidine kinase (HKs), also identified. Conversely and strangely, outer membrane protein gene ompW, unlike the gene ompC encoding receptor protein of T4 phage, was shown to provide phage resistance. Overall, this study exploited a genome-wide fitness assay to uncover susceptibility and resistant genes, even the shared genes, important for the E. coli strain of both most popular high lytic T4-like and T7-like phages. This knowledge of the genetic determinants can be further used to analysis the behind function signatures to screen the potential agents to aid phage killing of MDR pathogens, which will greatly be valuable in improving the phage therapy outcome in fighting with microbial resistance.
Keywords: Antimicrobial resistance (AMR); Lytic T4-like phage; Lytic T7-like phage; Tn5 transposons, Phage therapy.
Copyright © 2023 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare no conflict of interest.
Figures



Similar articles
-
Transposon Insertion Sequencing Elucidates Novel Gene Involvement in Susceptibility and Resistance to Phages T4 and T7 in Escherichia coli O157.mBio. 2018 Jul 24;9(4):e00705-18. doi: 10.1128/mBio.00705-18. mBio. 2018. PMID: 30042196 Free PMC article.
-
Construction of Leaderless-Bacteriocin-Producing Bacteriophage Targeting E. coli and Neighboring Gram-Positive Pathogens.Microbiol Spectr. 2021 Sep 3;9(1):e0014121. doi: 10.1128/Spectrum.00141-21. Epub 2021 Jul 14. Microbiol Spectr. 2021. PMID: 34259542 Free PMC article.
-
Manipulating Interactions between T4 Phage Long Tail Fibers and Escherichia coli Receptors.Appl Environ Microbiol. 2021 Jun 11;87(13):e0042321. doi: 10.1128/AEM.00423-21. Epub 2021 Jun 11. Appl Environ Microbiol. 2021. PMID: 33893116 Free PMC article.
-
Xenogeneic Regulation of the Bacterial Transcription Machinery.J Mol Biol. 2019 Sep 20;431(20):4078-4092. doi: 10.1016/j.jmb.2019.02.008. Epub 2019 Feb 15. J Mol Biol. 2019. PMID: 30776429 Review.
-
Advances in the T7 phage display system (Review).Mol Med Rep. 2018 Jan;17(1):714-720. doi: 10.3892/mmr.2017.7994. Epub 2017 Nov 7. Mol Med Rep. 2018. PMID: 29115536 Review.
Cited by
-
Coevolutionary phage training and Joint application delays the emergence of phage resistance in Pseudomonas aeruginosa.Virus Evol. 2023 Nov 18;9(2):vead067. doi: 10.1093/ve/vead067. eCollection 2023. Virus Evol. 2023. PMID: 38089014 Free PMC article.
-
An effective antibiofilm strategy based on bacteriophages armed with silver nanoparticles.Sci Rep. 2024 Apr 20;14(1):9088. doi: 10.1038/s41598-024-59866-y. Sci Rep. 2024. PMID: 38643290 Free PMC article.
-
Microfluidic Ecology Unravels the Genetic and Ecological Drivers of T4r Bacteriophage Resistance in E. coli: Insights into Biofilm-Mediated Evolution.Res Sq [Preprint]. 2024 May 24:rs.3.rs-4356333. doi: 10.21203/rs.3.rs-4356333/v1. Res Sq. 2024. PMID: 38826273 Free PMC article. Preprint.
-
Armed Phages: A New Weapon in the Battle Against Antimicrobial Resistance.Viruses. 2025 Jun 27;17(7):911. doi: 10.3390/v17070911. Viruses. 2025. PMID: 40733529 Free PMC article. Review.
References
-
- Artola-Recolons C., Lee M., Bernardo-García N., Blázquez B., Hesek D., Bartual S.G., Mahasenan K.V., Lastochkin E., Pi H., Boggess B., Meindl K., Usón I., Fisher J.F., Mobashery S., Hermoso J.A. Structure and cell wall cleavage by modular lytic transglycosylase MltC of Escherichia coli. ACS Chem. Biol. 2014;9(9):2058–2066. - PMC - PubMed
-
- Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., Pyshkin A.V., Sirotkin A.V., Vyahhi N., Tesler G., Alekseyev M.A., Pevzner P.A. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19(5):455–477. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

