SPRTN patient variants cause global-genome DNA-protein crosslink repair defects
- PMID: 36681662
- PMCID: PMC9867749
- DOI: 10.1038/s41467-023-35988-1
SPRTN patient variants cause global-genome DNA-protein crosslink repair defects
Abstract
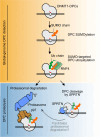
DNA-protein crosslinks (DPCs) are pervasive DNA lesions that are induced by reactive metabolites and various chemotherapeutic agents. Here, we develop a technique for the Purification of x-linked Proteins (PxP), which allows identification and tracking of diverse DPCs in mammalian cells. Using PxP, we investigate DPC repair in cells genetically-engineered to express variants of the SPRTN protease that cause premature ageing and early-onset liver cancer in Ruijs-Aalfs syndrome patients. We find an unexpected role for SPRTN in global-genome DPC repair, that does not rely on replication-coupled detection of the lesion. Mechanistically, we demonstrate that replication-independent DPC cleavage by SPRTN requires SUMO-targeted ubiquitylation of the protein adduct and occurs in addition to proteasomal DPC degradation. Defective ubiquitin binding of SPRTN patient variants compromises global-genome DPC repair and causes synthetic lethality in combination with a reduction in proteasomal DPC repair capacity.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

