Circular RNA circGlis3 protects against islet β-cell dysfunction and apoptosis in obesity
- PMID: 36681689
- PMCID: PMC9867769
- DOI: 10.1038/s41467-023-35998-z
Circular RNA circGlis3 protects against islet β-cell dysfunction and apoptosis in obesity
Abstract
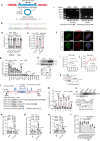
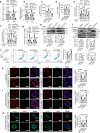
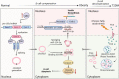
Pancreatic β-cell compensation is a major mechanism in delaying T2DM progression. Here we report the abnormal high expression of circGlis3 in islets of male mice with obesity and serum of people with obesity. Increasing circGlis3 is regulated by Quaking (QKI)-mediated splicing circularization. circGlis3 overexpression enhances insulin secretion and inhibits obesity-induced apoptosis in vitro and in vivo. Mechanistically, circGlis3 promotes insulin secretion by up-regulating NeuroD1 and Creb1 via sponging miR-124-3p and decreases apoptosis via interacting with the pro-apoptotic factor SCOTIN. The RNA binding protein FUS recruits circGlis3 and collectively assemble abnormal stable cytoplasmic stress granules (SG) in response to cellular stress. These findings highlight a physiological role for circRNAs in β-cell compensation and indicate that modulation of circGlis3 expression may represent a potential strategy to prevent β-cell dysfunction and apoptosis after obesity.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Lipotoxicity-induced circGlis3 impairs beta cell function and is transmitted by exosomes to promote islet endothelial cell dysfunction.Diabetologia. 2022 Jan;65(1):188-205. doi: 10.1007/s00125-021-05591-4. Epub 2021 Nov 9. Diabetologia. 2022. PMID: 34751796
-
A circular RNA derived from GLIS3 accelerates the proliferation of glioblastoma cells through competitively binding with miR-449c-5p to upregulate CAPG and GLIS3.BMC Neurosci. 2022 Sep 16;23(1):53. doi: 10.1186/s12868-022-00736-6. BMC Neurosci. 2022. PMID: 36114444 Free PMC article.
-
The long non-coding RNA βFaar regulates islet β-cell function and survival during obesity in mice.Nat Commun. 2021 Jun 28;12(1):3997. doi: 10.1038/s41467-021-24302-6. Nat Commun. 2021. PMID: 34183666 Free PMC article.
-
Circular RNAs as Novel Regulators of β-Cell Functions under Physiological and Pathological Conditions.Int J Mol Sci. 2021 Feb 3;22(4):1503. doi: 10.3390/ijms22041503. Int J Mol Sci. 2021. PMID: 33546109 Free PMC article. Review.
-
An 'alpha-beta' of pancreatic islet microribonucleotides.Int J Biochem Cell Biol. 2017 Jul;88:208-219. doi: 10.1016/j.biocel.2017.01.009. Epub 2017 Jan 22. Int J Biochem Cell Biol. 2017. PMID: 28122254 Review.
Cited by
-
EIF4A3-Mediated Biogenesis of CircFADS1 Promotes the Progression of Hepatocellular Carcinoma via Wnt/β-Catenin Pathway.Adv Sci (Weinh). 2025 Apr;12(14):e2411869. doi: 10.1002/advs.202411869. Epub 2025 Feb 18. Adv Sci (Weinh). 2025. PMID: 39965082 Free PMC article.
-
Homocysteine Promotes the Pathogenesis of Atherosclerosis through the Circ-PIAS1-5/miR-219a-2-3p/TEAD1 Axis.Adv Sci (Weinh). 2025 May;12(18):e2415563. doi: 10.1002/advs.202415563. Epub 2025 Mar 16. Adv Sci (Weinh). 2025. PMID: 40089857 Free PMC article.
-
Escherichia coli-Induced cGLIS3-Mediated Stress Granules Activate the NF-κB Pathway to Promote Intrahepatic Cholangiocarcinoma Progression.Adv Sci (Weinh). 2024 Apr;11(16):e2306174. doi: 10.1002/advs.202306174. Epub 2024 Feb 17. Adv Sci (Weinh). 2024. PMID: 38368261 Free PMC article.
-
RNA-binding protein quaking: a multifunctional regulator in tumour progression.Ann Med. 2025 Dec;57(1):2443046. doi: 10.1080/07853890.2024.2443046. Epub 2024 Dec 23. Ann Med. 2025. PMID: 39711373 Free PMC article. Review.
-
miR-423 sponged by lncRNA NORHA inhibits granulosa cell apoptosis.J Anim Sci Biotechnol. 2023 Dec 5;14(1):154. doi: 10.1186/s40104-023-00960-y. J Anim Sci Biotechnol. 2023. PMID: 38053184 Free PMC article.
References
-
- James, E. et al. NCD Countdown 2030: worldwide trends in non-communicable disease mortality and progress towards Sustainable Development Goal target 3.4. Lancet392, 1072–1088 (2018). - PubMed
-
- Butler AE, Janson J, Soeller WC, Butler PC. Increased beta-cell apoptosis prevents adaptive increase in beta-cell mass in mouse model of type 2 diabetes: evidence for role of islet amyloid formation rather than direct action of amyloid. Diabetes. 2003;52:2304–2314. doi: 10.2337/diabetes.52.9.2304. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

