Genomic landscape of the emerging XDR Salmonella Typhi for mining druggable targets clpP, hisH, folP and gpmI and screening of novel TCM inhibitors, molecular docking and simulation analyses
- PMID: 36681806
- PMCID: PMC9860245
- DOI: 10.1186/s12866-023-02756-6
Genomic landscape of the emerging XDR Salmonella Typhi for mining druggable targets clpP, hisH, folP and gpmI and screening of novel TCM inhibitors, molecular docking and simulation analyses
Abstract
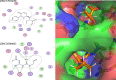
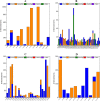
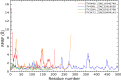
Typhoid fever is transmitted by ingestion of polluted water, contaminated food, and stool of typhoid-infected individuals, mostly in developing countries with poor hygienic environments. To find novel therapeutic targets and inhibitors, We employed a subtractive genomics strategy towards Salmonella Typhi and the complete genomes of eight strains were primarily subjected to the EDGAR tool to predict the core genome (n = 3207). Human non-homology (n = 2450) was followed by essential genes identification (n = 37). The STRING database predicted maximum protein-protein interactions, followed by cellular localization. The virulent/immunogenic ability of predicted genes were checked to differentiate drug and vaccine targets. Furthermore, the 3D models of the identified putative proteins encoded by the respective genes were constructed and subjected to druggability analyses where only "highly druggable" proteins were selected for molecular docking and simulation analyses. The putative targets ATP-dependent CLP protease proteolytic subunit, Imidazole glycerol phosphate synthase hisH, 7,8-dihydropteroate synthase folP and 2,3-bisphosphoglycerate-independent phosphoglycerate mutase gpmI were screened against a drug-like library (n = 12,000) and top hits were selected based on H-bonds, RMSD and energy scores. Finally, the ADMET properties for novel inhibitors ZINC19340748, ZINC09319798, ZINC00494142, ZINC32918650 were optimized followed by binding free energy (MM/PBSA) calculation for ligand-receptor complexes. The findings of this work are expected to aid in expediting the identification of novel protein targets and inhibitors in combating typhoid Salmonellosis, in addition to the already existing therapies.
Keywords: MD simulation; Salmonella Typhi; Screening and ADMET profiling; Subtractive genomics.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures





Similar articles
-
Comparative genomics study for identification of putative drug targets in Salmonella typhi Ty2.Gene. 2016 Jan 15;576(1 Pt 3):544-59. doi: 10.1016/j.gene.2015.11.007. Epub 2015 Nov 10. Gene. 2016. PMID: 26555890
-
Identification of a Novel Therapeutic Target against XDR Salmonella Typhi H58 Using Genomics Driven Approach Followed Up by Natural Products Virtual Screening.Microorganisms. 2021 Dec 3;9(12):2512. doi: 10.3390/microorganisms9122512. Microorganisms. 2021. PMID: 34946114 Free PMC article.
-
Nanoparticle Fullerene (C60) demonstrated stable binding with antibacterial potential towards probable targets of drug resistant Salmonella typhi - a computational perspective and in vitro investigation.J Biomol Struct Dyn. 2017 Dec;35(16):3449-3468. doi: 10.1080/07391102.2016.1257441. Epub 2016 Nov 23. J Biomol Struct Dyn. 2017. PMID: 27817242
-
Extensively Drug-Resistant (XDR) Typhoid: Evolution, Prevention, and Its Management.Biomed Res Int. 2020 May 2;2020:6432580. doi: 10.1155/2020/6432580. eCollection 2020. Biomed Res Int. 2020. PMID: 32462008 Free PMC article. Review.
-
Salmonella Typhi genomics: envisaging the future of typhoid eradication.Trop Med Int Health. 2017 Aug;22(8):918-925. doi: 10.1111/tmi.12899. Epub 2017 Jun 19. Trop Med Int Health. 2017. PMID: 28544285 Review.
Cited by
-
Exploring Nocardia's ecological spectrum and novel therapeutic frontiers through whole-genome sequencing: unraveling drug resistance and virulence factors.Arch Microbiol. 2024 Jan 24;206(2):76. doi: 10.1007/s00203-023-03799-z. Arch Microbiol. 2024. PMID: 38267747
-
Bridging drug discovery through hierarchical subtractive genomics against asd, trpG, and secY of pneumonia causing MDR Staphylococcus aureus.Mol Genet Genomics. 2024 Mar 13;299(1):34. doi: 10.1007/s00438-024-02115-8. Mol Genet Genomics. 2024. PMID: 38478130
-
Computational Strategies in Drug Discovery: Leveraging Subtractive Genomic Analysis for Target Identification.Curr Pharm Biotechnol. 2025;26(9):1271-1286. doi: 10.2174/0113892010295441240515074348. Curr Pharm Biotechnol. 2025. PMID: 38808712 Review.
-
Structure-Activity Relationship Target Prediction Studies of Clindamycin Derivatives with Broad-Spectrum Bacteriostatic Antibacterial Properties.Molecules. 2023 Oct 31;28(21):7357. doi: 10.3390/molecules28217357. Molecules. 2023. PMID: 37959776 Free PMC article.
-
Druggable proteins of Alistipes reveal promising antimicrobial targets against chronic intestinal inflammation.Front Pharmacol. 2025 Apr 24;16:1555062. doi: 10.3389/fphar.2025.1555062. eCollection 2025. Front Pharmacol. 2025. PMID: 40342998 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

