Identification of a short ACE2-derived stapled peptide targeting the SARS-CoV-2 spike protein
- PMID: 36682293
- PMCID: PMC9842534
- DOI: 10.1016/j.ejmech.2023.115118
Identification of a short ACE2-derived stapled peptide targeting the SARS-CoV-2 spike protein
Abstract
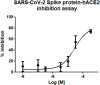
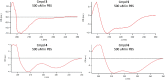
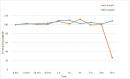
The design and synthesis of a series of peptide derivatives based on a short ACE2 α-helix 1 epitope and subsequent [i - i+4] stapling of the secondary structure resulted in the identification of a 9-mer peptide capable to compete with recombinant ACE2 towards Spike RBD in the micromolar range. Specifically, SARS-CoV-2 Spike inhibitor screening based on colorimetric ELISA assay and structural studies by circular dichroism showed the ring-closing metathesis cyclization being capable to stabilize the helical structure of the 9-mer 34HEAEDLFYQ42 epitope better than the triazole stapling via click chemistry. MD simulations showed the stapled peptide being able not only to bind the Spike RBD, sterically interfering with ACE2, but also showing higher affinity to the target as compared to parent epitope.
Keywords: COVID-19; Infectious diseases; Molecular modeling; Peptidomimetics; Protein-protein interaction; Ring-closing metathesis.
Copyright © 2023 Elsevier Masson SAS. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
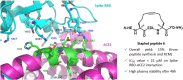
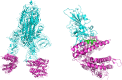
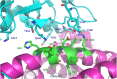

References
-
- WHO, Coronavirus disease (COVID-19) pandemic. https://www.who.int/emergencies/diseases/novel-coronavirus-2019
-
- Worldometer, Cases Coronavirus. https://www.worldometers.info/coronavirus/coronavirus-cases/#daily-cases
-
- Callaway E. Beyond Omicron: what's next for COVID's viral evolution. Nature. 2021;600:204–207. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

