Alternative 3'-untranslated regions regulate high-salt tolerance of Spartina alterniflora
- PMID: 36682816
- PMCID: PMC10069910
- DOI: 10.1093/plphys/kiad030
Alternative 3'-untranslated regions regulate high-salt tolerance of Spartina alterniflora
Erratum in
-
Correction to: Alternative 3'-untranslated regions regulate high-salt tolerance of Spartina alterniflora.Plant Physiol. 2023 Jul 3;192(3):2569. doi: 10.1093/plphys/kiad211. Plant Physiol. 2023. PMID: 37022949 Free PMC article. No abstract available.
Abstract
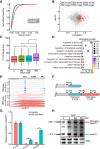
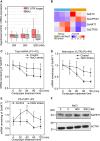
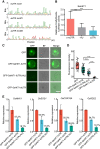
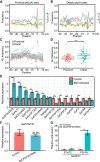
High-salt stress continues to challenge the growth and survival of many plants. Alternative polyadenylation (APA) produces mRNAs with different 3'-untranslated regions (3' UTRs) to regulate gene expression at the post-transcriptional level. However, the roles of alternative 3' UTRs in response to salt stress remain elusive. Here, we report the function of alternative 3' UTRs in response to high-salt stress in S. alterniflora (Spartina alterniflora), a monocotyledonous halophyte tolerant of high-salt environments. We found that high-salt stress induced global APA dynamics, and ∼42% of APA genes responded to salt stress. High-salt stress led to 3' UTR lengthening of 207 transcripts through increasing the usage of distal poly(A) sites. Transcripts with alternative 3' UTRs were mainly enriched in salt stress-related ion transporters. Alternative 3' UTRs of HIGH-AFFINITY K+ TRANSPORTER 1 (SaHKT1) increased RNA stability and protein synthesis in vivo. Regulatory AU-rich elements were identified in alternative 3' UTRs, boosting the protein level of SaHKT1. RNAi-knock-down experiments revealed that the biogenesis of 3' UTR lengthening in SaHKT1 was controlled by the poly(A) factor CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR 30 (SaCPSF30). Over-expression of SaHKT1 with an alternative 3' UTR in rice (Oryza sativa) protoplasts increased mRNA accumulation of salt-tolerance genes in an AU-rich element-dependent manner. These results suggest that mRNA 3' UTR lengthening is a potential mechanism in response to high-salt stress. These results also reveal complex regulatory roles of alternative 3' UTRs coupling APA and regulatory elements at the post-transcriptional level in plants.
© American Society of Plant Biologists 2023. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Conflict of interest statement
Conflict of interest statement. None declared.
Figures



Similar articles
-
The Full-Length Transcriptome of Spartina alterniflora Reveals the Complexity of High Salt Tolerance in Monocotyledonous Halophyte.Plant Cell Physiol. 2020 May 1;61(5):882-896. doi: 10.1093/pcp/pcaa013. Plant Cell Physiol. 2020. PMID: 32044993
-
The Spartina alterniflora genome sequence provides insights into the salt-tolerance mechanisms of exo-recretohalophytes.Plant Biotechnol J. 2024 Sep;22(9):2558-2574. doi: 10.1111/pbi.14368. Epub 2024 Apr 29. Plant Biotechnol J. 2024. PMID: 38685729 Free PMC article.
-
Deep learning-enabled discovery and characterization of HKT genes in Spartina alterniflora.Plant J. 2023 Nov;116(3):690-705. doi: 10.1111/tpj.16397. Epub 2023 Jul 26. Plant J. 2023. PMID: 37494542
-
Implications of polyadenylation in health and disease.Nucleus. 2014;5(6):508-19. doi: 10.4161/nucl.36360. Epub 2014 Oct 31. Nucleus. 2014. PMID: 25484187 Free PMC article. Review.
-
Emerging Roles for 3' UTRs in Neurons.Int J Mol Sci. 2020 May 12;21(10):3413. doi: 10.3390/ijms21103413. Int J Mol Sci. 2020. PMID: 32408514 Free PMC article. Review.
Cited by
-
Dynamics of alternative polyadenylation in single root cells of Arabidopsis thaliana.Front Plant Sci. 2024 Sep 20;15:1437118. doi: 10.3389/fpls.2024.1437118. eCollection 2024. Front Plant Sci. 2024. PMID: 39372861 Free PMC article.
-
Genome-wide characterization of post-transcriptional processes related to wood formation in Dalbergia odorifera.BMC Genomics. 2024 Apr 16;25(1):372. doi: 10.1186/s12864-024-10300-7. BMC Genomics. 2024. PMID: 38627613 Free PMC article.
-
Functional Consequences of Shifting Transcript Boundaries in Glucose Starvation.Mol Cell Biol. 2023;43(11):611-628. doi: 10.1080/10985549.2023.2270406. Epub 2023 Nov 17. Mol Cell Biol. 2023. PMID: 37937348 Free PMC article.
-
AAAP gene family evolution and transcriptional regulation in Eucalyptus grandis under nitrogen, phosphate and boron deficiencies.BMC Plant Biol. 2025 Jul 4;25(1):879. doi: 10.1186/s12870-025-06907-x. BMC Plant Biol. 2025. PMID: 40615974 Free PMC article.
-
Decoding PHR-Orchestrated Stress Adaptation: A Genome-Wide Integrative Analysis of Transcriptional Regulation Under Abiotic Stress in Eucalyptus grandis.Int J Mol Sci. 2025 Mar 25;26(7):2958. doi: 10.3390/ijms26072958. Int J Mol Sci. 2025. PMID: 40243569 Free PMC article.
References
-
- Chen C-Y, Gherzi R, Ong S-E, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M (2001) AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107(4): 451–464 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

