Comparative and expression analyses of AP2/ERF genes reveal copy number expansion and potential functions of ERF genes in Solanaceae
- PMID: 36683040
- PMCID: PMC9869560
- DOI: 10.1186/s12870-022-04017-6
Comparative and expression analyses of AP2/ERF genes reveal copy number expansion and potential functions of ERF genes in Solanaceae
Abstract
Background: The AP2/ERF gene family is a superfamily of transcription factors that are important in the response of plants to abiotic stress and development. However, comprehensive research of the AP2/ERF genes in the Solanaceae family is lacking.
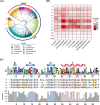
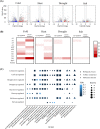
Results: Here, we updated the annotation of AP2/ERF genes in the genomes of eight Solanaceae species, as well as Arabidopsis thaliana and Oryza sativa. We identified 2,195 AP2/ERF genes, of which 368 (17%) were newly identified. Based on phylogenetic analyses, we observed expansion of the copy number of these genes, especially those belonging to specific Ethylene-Responsive Factor (ERF) subgroups of the Solanaceae. From the results of chromosomal location and synteny analyses, we identified that the AP2/ERF genes of the pepper (Capsicum annuum), the tomato (Solanum lycopersicum), and the potato (Solanum tuberosum) belonging to ERF subgroups form a tandem array and most of them are species-specific without orthologs in other species, which has led to differentiation of AP2/ERF gene repertory among Solanaceae. We suggest that these genes mainly emerged through recent gene duplication after the divergence of these species. Transcriptome analyses showed that the genes have a putative function in the response of the pepper and tomato to abiotic stress, especially those in ERF subgroups.
Conclusions: Our findings will provide comprehensive information on AP2/ERF genes and insights into the structural, evolutionary, and functional understanding of the role of these genes in the Solanaceae.
Keywords: AP2/ERF; Abiotic stress; Re-annotation; Solanaceae; Transcription factors; species-specific duplication.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Bilas R, Szafran K, Hnatuszko-Konka K, Kononowicz AK. Cis-regulatory elements used to control gene expression in plants. Plant Cell Tiss Org. 2016;127(2):269–287. doi: 10.1007/s11240-016-1057-7. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

