De novo PAM generation to reach initially inaccessible target sites for base editing
- PMID: 36683434
- PMCID: PMC10110497
- DOI: 10.1242/dev.201115
De novo PAM generation to reach initially inaccessible target sites for base editing
Abstract
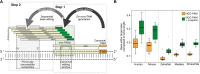
Base editing by CRISPR crucially depends on the presence of a protospacer adjacent motif (PAM) at the correct distance from the editing site. Here, we present and validate an efficient one-shot approach termed 'inception' that expands the editing range. This is achieved by sequential, combinatorial base editing: de novo generated synonymous, non-synonymous or intronic PAM sites facilitate subsequent base editing at nucleotide positions that were initially inaccessible, further opening the targeting range of highly precise editing approaches. We demonstrate the applicability of the inception concept in medaka (Oryzias latipes) in three settings: loss of function, by introducing a pre-termination STOP codon in the open reading frame of oca2; locally confined multi-codon changes to generate allelic variants with different phenotypic severity in kcnh6a; and the removal of a splice acceptor site by targeting intronic sequences of rx3. Using sequentially acting base editors in the described combinatorial approach expands the number of accessible target sites by 65% on average. This allows the use of well-established tools with NGG PAM recognition for the establishment of thus far unreachable disease models, for hypomorphic allele studies and for efficient targeted mechanistic investigations in a precise and predictable manner.
Keywords: De novo PAM; Base editing; CRISPR; Medaka; Targeted mutagenesis.
© 2023. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures



References
-
- García-Tuñón, I., Alonso-Pérez, V., Vuelta, E., Pérez-Ramos, S., Herrero, M., Méndez, L., Hernández-Sánchez, J. M., Martín-Izquierdo, M., Sáldana, R., Sevilla, J.et al. (2019). Splice donor site sgRNAs enhance CRISPR/Cas9-mediated knockout efficiency. PLoS One 14, e0216674. 10.1371/journal.pone.0216674 - DOI - PMC - PubMed
-
- Hadley, W. (2016). Ggplot2. New York, NY: Springer Science+Business Media, LLC.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

