Rapid detection of influenza A viruses using a real-time reverse transcription recombinase-aided amplification assay
- PMID: 36683681
- PMCID: PMC9849684
- DOI: 10.3389/fcimb.2022.1071288
Rapid detection of influenza A viruses using a real-time reverse transcription recombinase-aided amplification assay
Abstract
Introduction: Influenza A viruses (IAVs) are important pathogens of respiratory infections, causing not only seasonal influenza but also influenza pandemics and posing a global threat to public health. IAVs infection spreads rapidly, widely, and across species, causing huge losses, especially zoonotic IAVs infections that are more harmful. Fast and sensitive detection of IAVs is critical for controlling the spread of this disease.
Methods: Here, a real-time reverse transcription recombinase-aided amplification (real-time RT-RAA) assay targeting conserved positions in the matrix protein gene (M gene) of IAVs, is successfully established to detect IAVs. The assay can be completed within 20 min at 42°C.
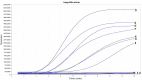
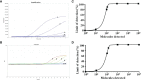
Results: The sensitivity of the real-time RT-RAA assay was 142 copies per reaction at 95% probability, which was comparable to the sensitivity of the RT-qPCR assay. The specificity assay showed that the real-time RT-RAA assay was specific to IAVs, and there was no cross-reactivity with other important viruses. In addition, 100%concordance between the real-time RT-RAA and RT-qPCR assays was achieved after testing 120 clinical specimens.
Discussion: The results suggested that the real-time RT-RAA assay we developed was a specific, sensitive and reliable diagnostic tool for the rapid detection of IAVs.
Keywords: clinical diagnosis; influenza A viruses (IAVs); isothermal amplification; rapid diagnosis; reverse transcription recombinase-aided amplification (RT-RAA).
Copyright © 2023 Cui, Zhang, Tu, Zhao, Kong, Pu, Zhang, Chen, Sun, Wei, Liang, Liu, Liu and Guo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

