Emergence of plasmid-mediated colistin resistance mcr-3.5 gene in Citrobacter amalonaticus and Citrobacter sedlakii isolated from healthy individual in Thailand
- PMID: 36683683
- PMCID: PMC9846275
- DOI: 10.3389/fcimb.2022.1067572
Emergence of plasmid-mediated colistin resistance mcr-3.5 gene in Citrobacter amalonaticus and Citrobacter sedlakii isolated from healthy individual in Thailand
Abstract
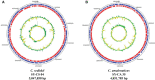
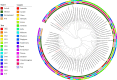
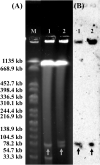
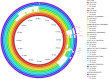
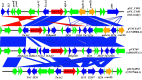
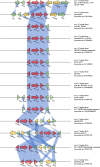
Citrobacter spp. are Gram-negative bacteria commonly found in environments and intestinal tracts of humans and animals. They are generally susceptible to third-generation cephalosporins, carbapenems and colistin. However, several antibiotic resistant genes have been increasingly reported in Citrobacter spp., which leads to the postulation that Citrobacter spp. could potentially be a reservoir for spreading of antimicrobial resistant genes. In this study, we characterized two colistin-resistant Citrobacter spp. isolated from the feces of a healthy individual in Thailand. Based on MALDI-TOF and ribosomal multilocus sequence typing, both strains were identified as Citrobacter sedlakii and Citrobacter amalonaticus. Genomic analysis and S1-nuclease pulsed field gel electrophoresis/DNA hybridization revealed that Citrobacter sedlakii and Citrobacter amalonaticus harbored mcr-3.5 gene on pSY_CS01 and pSY_CA01 plasmids, respectively. Both plasmids belonged to IncFII(pCoo) replicon type, contained the same genetic context (Tn3-IS1-ΔTnAs2-mcr-3.5-dgkA-IS91) and exhibited high transferring frequencies ranging from 1.03×10-4 - 4.6×10-4 CFU/recipient cell Escherichia coli J53. Colistin-MICs of transconjugants increased ≥ 16-fold suggesting that mcr-3.5 on these plasmids can be expressed in other species. However, beside mcr, other major antimicrobial resistant determinants in multidrug resistant Enterobacterales were not found in these two isolates. These findings indicate that mcr gene continued to evolve in the absence of antibiotics selective pressure. Our results also support the hypothesis that Citrobacter could be a reservoir for spreading of antimicrobial resistant genes. To the best of our knowledge, this is the first report that discovered human-derived Citrobacter spp. that harbored mcr but no other major antimicrobial resistant determinants. Also, this is the first report that described the presence of mcr gene in C. sedlakii and mcr-3 in C. amalonaticus.
Keywords: citrobacter amalonaticus; citrobacter sedlakii; citrobacter spp.; colistin resistance; mcr gene; mcr-3.
Copyright © 2023 Phuadraksa, Wichit, Songtawee, Tantimavanich, Isarankura-Na-Ayudhya and Yainoy.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Characterization of a carbapenem-resistant Citrobacter amalonaticus coharbouring bla IMP-4 and qnrs1 genes.J Med Microbiol. 2021 Jun;70(6). doi: 10.1099/jmm.0.001364. J Med Microbiol. 2021. PMID: 34170219
-
Isolation of an IncP-1 plasmid harbouring mcr-1 from a chicken isolate of Citrobacter braakii in China.Int J Antimicrob Agents. 2018 Jun;51(6):936-940. doi: 10.1016/j.ijantimicag.2017.12.030. Epub 2018 Jan 3. Int J Antimicrob Agents. 2018. PMID: 29305957
-
Colistin resistance and plasmid-mediated mcr genes in Escherichia coli and Salmonella isolated from pigs, pig carcass and pork in Thailand, Lao PDR and Cambodia border provinces.J Vet Sci. 2021 Sep;22(5):e68. doi: 10.4142/jvs.2021.22.e68. Epub 2021 Aug 3. J Vet Sci. 2021. PMID: 34423604 Free PMC article.
-
mcr Genes Conferring Colistin Resistance in Enterobacterales; a Five Year Overview.Acta Med Acad. 2021 Dec;50(3):365-371. doi: 10.5644/ama2006-124.355. Acta Med Acad. 2021. PMID: 35164512 Review.
-
Environmental mediation of colistin resistance in the African context. A systematic scoping review.J Glob Antimicrob Resist. 2025 Mar;41:39-43. doi: 10.1016/j.jgar.2024.12.002. Epub 2024 Dec 14. J Glob Antimicrob Resist. 2025. PMID: 39681218
Cited by
-
Mobile Colistin-Resistant Genes mcr-1, mcr-2, and mcr-3 Identified in Diarrheal Pathogens among Infants, Children, and Adults in Bangladesh: Implications for the Future.Antibiotics (Basel). 2024 Jun 7;13(6):534. doi: 10.3390/antibiotics13060534. Antibiotics (Basel). 2024. PMID: 38927200 Free PMC article.
-
A brief insight into Citrobacter species - a growing threat to public health.Front Antibiot. 2023 Dec 5;2:1276982. doi: 10.3389/frabi.2023.1276982. eCollection 2023. Front Antibiot. 2023. PMID: 39816660 Free PMC article. Review.
-
Trends in Colistin Resistance and Multidrug-Resistant Phenotypes Among Gram-Negative Bacilli: A Retrospective Analysis.Molecules. 2025 Jul 12;30(14):2950. doi: 10.3390/molecules30142950. Molecules. 2025. PMID: 40733216 Free PMC article.
-
Occurrence and mechanisms of tigecycline resistance in carbapenem- and colistin-resistant Klebsiella pneumoniae in Thailand.Sci Rep. 2024 Mar 3;14(1):5215. doi: 10.1038/s41598-024-55705-2. Sci Rep. 2024. PMID: 38433246 Free PMC article.
-
The Gut Microbiome of the Asiatic Toad (Bufo gargarizans) Reflects Environmental Changes and Human Activities.Ecol Evol. 2025 May 7;15(5):e71394. doi: 10.1002/ece3.71394. eCollection 2025 May. Ecol Evol. 2025. PMID: 40342698 Free PMC article.
References
-
- AbuOun M., Stubberfield E. J., Duggett N. A., Kirchner M., Dormer L., Nunez-Garcia J., et al. (2018). Mcr-1 and mcr-2 (mcr-6.1) variant genes identified in moraxella species isolated from pigs in great Britain from 2014 to 2015. J. Antimicrob. Chemother. 73, 2904. doi: 10.1093/jac/dky272 - DOI - PMC - PubMed
-
- Andrews S. FastQC: A quality control tool for high throughput sequence data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (Accessed February 2, 2022).
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

