Emergence of plasmid-mediated colistin resistance mcr-3.5 gene in Citrobacter amalonaticus and Citrobacter sedlakii isolated from healthy individual in Thailand
- PMID: 36683683
- PMCID: PMC9846275
- DOI: 10.3389/fcimb.2022.1067572
Emergence of plasmid-mediated colistin resistance mcr-3.5 gene in Citrobacter amalonaticus and Citrobacter sedlakii isolated from healthy individual in Thailand
Abstract
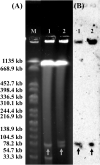
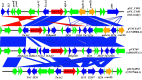
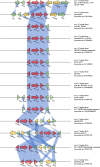
Citrobacter spp. are Gram-negative bacteria commonly found in environments and intestinal tracts of humans and animals. They are generally susceptible to third-generation cephalosporins, carbapenems and colistin. However, several antibiotic resistant genes have been increasingly reported in Citrobacter spp., which leads to the postulation that Citrobacter spp. could potentially be a reservoir for spreading of antimicrobial resistant genes. In this study, we characterized two colistin-resistant Citrobacter spp. isolated from the feces of a healthy individual in Thailand. Based on MALDI-TOF and ribosomal multilocus sequence typing, both strains were identified as Citrobacter sedlakii and Citrobacter amalonaticus. Genomic analysis and S1-nuclease pulsed field gel electrophoresis/DNA hybridization revealed that Citrobacter sedlakii and Citrobacter amalonaticus harbored mcr-3.5 gene on pSY_CS01 and pSY_CA01 plasmids, respectively. Both plasmids belonged to IncFII(pCoo) replicon type, contained the same genetic context (Tn3-IS1-ΔTnAs2-mcr-3.5-dgkA-IS91) and exhibited high transferring frequencies ranging from 1.03×10-4 - 4.6×10-4 CFU/recipient cell Escherichia coli J53. Colistin-MICs of transconjugants increased ≥ 16-fold suggesting that mcr-3.5 on these plasmids can be expressed in other species. However, beside mcr, other major antimicrobial resistant determinants in multidrug resistant Enterobacterales were not found in these two isolates. These findings indicate that mcr gene continued to evolve in the absence of antibiotics selective pressure. Our results also support the hypothesis that Citrobacter could be a reservoir for spreading of antimicrobial resistant genes. To the best of our knowledge, this is the first report that discovered human-derived Citrobacter spp. that harbored mcr but no other major antimicrobial resistant determinants. Also, this is the first report that described the presence of mcr gene in C. sedlakii and mcr-3 in C. amalonaticus.
Keywords: citrobacter amalonaticus; citrobacter sedlakii; citrobacter spp.; colistin resistance; mcr gene; mcr-3.
Copyright © 2023 Phuadraksa, Wichit, Songtawee, Tantimavanich, Isarankura-Na-Ayudhya and Yainoy.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- AbuOun M., Stubberfield E. J., Duggett N. A., Kirchner M., Dormer L., Nunez-Garcia J., et al. (2018). Mcr-1 and mcr-2 (mcr-6.1) variant genes identified in moraxella species isolated from pigs in great Britain from 2014 to 2015. J. Antimicrob. Chemother. 73, 2904. doi: 10.1093/jac/dky272 - DOI - PMC - PubMed
-
- Andrews S. FastQC: A quality control tool for high throughput sequence data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (Accessed February 2, 2022).
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

