Splice-switch oligonucleotide-based combinatorial platform prioritizes synthetic lethal targets CHK1 and BRD4 against MYC-driven hepatocellular carcinoma
- PMID: 36684069
- PMCID: PMC9842033
- DOI: 10.1002/btm2.10363
Splice-switch oligonucleotide-based combinatorial platform prioritizes synthetic lethal targets CHK1 and BRD4 against MYC-driven hepatocellular carcinoma
Abstract
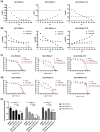
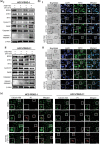
Deregulation of MYC is among the most frequent oncogenic drivers in hepatocellular carcinoma (HCC). Unfortunately, the clinical success of MYC-targeted therapies is limited. Synthetic lethality offers an alternative therapeutic strategy by leveraging on vulnerabilities in tumors with MYC deregulation. While several synthetic lethal targets of MYC have been identified in HCC, the need to prioritize targets with the greatest therapeutic potential has been unmet. Here, we demonstrate that by pairing splice-switch oligonucleotide (SSO) technologies with our phenotypic-analytical hybrid multidrug interrogation platform, quadratic phenotypic optimization platform (QPOP), we can disrupt the functional expression of these targets in specific combinatorial tests to rapidly determine target-target interactions and rank synthetic lethality targets. Our SSO-QPOP analyses revealed that simultaneous attenuation of CHK1 and BRD4 function is an effective combination specific in MYC-deregulated HCC, successfully suppressing HCC progression in vitro. Pharmacological inhibitors of CHK1 and BRD4 further demonstrated its translational value by exhibiting synergistic interactions in patient-derived xenograft organoid models of HCC harboring high levels of MYC deregulation. Collectively, our work demonstrates the capacity of SSO-QPOP as a target prioritization tool in the drug development pipeline, as well as the therapeutic potential of CHK1 and BRD4 in MYC-driven HCC.
Keywords: MYC synthetic lethality; RNA therapeutics; quadratic phenotypic optimization platform; splice‐switch oligonucleotides.
© 2022 The Authors. Bioengineering & Translational Medicine published by Wiley Periodicals LLC on behalf of American Institute of Chemical Engineers.
Conflict of interest statement
Edward Kai‐Hua Chow is a shareholder in KYAN Therapeutics.
Figures
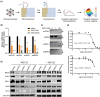
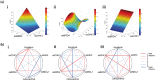
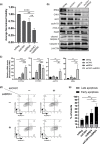
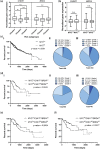
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

