Chromosome architecture and homologous recombination in meiosis
- PMID: 36684419
- PMCID: PMC9853400
- DOI: 10.3389/fcell.2022.1097446
Chromosome architecture and homologous recombination in meiosis
Abstract
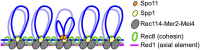
Meiocytes organize higher-order chromosome structures comprising arrays of chromatin loops organized at their bases by linear axes. As meiotic prophase progresses, the axes of homologous chromosomes align and synapse along their lengths to form ladder-like structures called synaptonemal complexes (SCs). The entire process of meiotic recombination, from initiation via programmed DNA double-strand breaks (DSBs) to completion of DSB repair with crossover or non-crossover outcomes, occurs in the context of chromosome axes and SCs. These meiosis-specific chromosome structures provide specialized environments for the regulation of DSB formation and crossing over. In this review, we summarize insights into the importance of chromosome architecture in the regulation of meiotic recombination, focusing on cohesin-mediated axis formation, DSB regulation via tethered loop-axis complexes, inter-homolog template bias facilitated by axial proteins, and crossover regulation in the context of the SCs. We also discuss emerging evidence that the SUMO and the ubiquitin-proteasome system function in the organization of chromosome structure and regulation of meiotic recombination.
Keywords: axis-loop structure; cohesin; crossover; meiotic recombination; synaptonemal complex.
Copyright © 2023 Ito and Shinohara.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
Publication types
LinkOut - more resources
Full Text Sources
Research Materials

