Differential expression of lysine acetylation proteins in gastric cancer treated with a new antitumor agent bioactive peptide chelate selenium
- PMID: 36684675
- PMCID: PMC9854375
- DOI: 10.7717/peerj.14384
Differential expression of lysine acetylation proteins in gastric cancer treated with a new antitumor agent bioactive peptide chelate selenium
Abstract
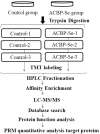
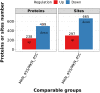
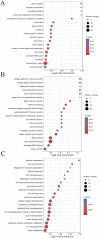
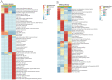
The method of anticancer bioactive peptide (ACBP) functionalized selenium particle (Se), which has enhanced anticancer activity, inhibited the growth of gastric cancer (GC) cells, and increased the ability of apoptosis in vitro, has been reported in previous studies. We used tandem mass spectrometry (TMT) labeling to construct a complete atlas of the acetylation-modified proteome in GC MKN-45 cells treated with ACBP-Se. The proteomics data database was searched and analyzed by bioinformatics: Kyoto Encyclopedia of Genes and Genomes (KEGG), Gene Ontology (GO), functional enrichment, and protein-protein interaction network. Finally, we conducted a quantitative PRM analysis of the selected target-modified peptides. We identified 4,958 acetylation sites from 1,926 proteins in this research. Among these, 4,467 acetylation sites corresponding to 1,777 proteins were quantified. Based on the above data and standards, we found that in the ACBP-Se group vs. the control group, 297 sites were upregulated, and 665 sites were downregulated. We systematically assessed the proteins containing quantitative information sites, including protein annotation, functional classification, and functional enrichment, cluster analysis supported by functional enrichment, domain structures, and protein interaction networks. Finally, we evaluated differentially expressed lysine acetylation sites. We revealed that SHMT2 K200 and PGK1 K97 were the most critical acetylated non-histone proteins, which may have an essential role in ACBP-Se treatment. Here, we identified and quantified the lysine acetylation proteins in GC cells treated with ACBP-Se. The characterization of acetylation indicates that acetylated proteins might be pivotal in the biological process, molecular binding, and metabolic pathways of ACBP-Se treatment progress. Our findings provide a broad understanding of acetylation ACBP-Se treatment of GC, suggesting a potential application for molecular targeted therapy.
Keywords: Acetylation; Anticancer bioactive peptides; Gastric cancer; Selenium.
©2023 Xu et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures





Similar articles
-
Global-scale profiling of differential expressed lysine acetylated proteins in colorectal cancer tumors and paired liver metastases.J Proteomics. 2016 Jun 16;142:24-32. doi: 10.1016/j.jprot.2016.05.002. Epub 2016 May 10. J Proteomics. 2016. PMID: 27178108
-
iTRAQ‑based proteomics analysis of the therapeutic effects of combined anticancer bioactive peptides and oxaliplatin on gastric cancer cells.Oncol Rep. 2020 Jan;43(1):201-217. doi: 10.3892/or.2019.7406. Epub 2019 Nov 11. Oncol Rep. 2020. PMID: 31746436 Free PMC article.
-
Antioxidant stress and anticancer activity of peptide‑chelated selenium in vitro.Int J Mol Med. 2021 Aug;48(2):153. doi: 10.3892/ijmm.2021.4986. Epub 2021 Jun 24. Int J Mol Med. 2021. PMID: 34165159 Free PMC article.
-
Anticancer bioactive peptide-3 inhibits human gastric cancer growth by suppressing gastric cancer stem cells.J Cell Biochem. 2014 Apr;115(4):697-711. doi: 10.1002/jcb.24711. J Cell Biochem. 2014. PMID: 24214799
-
Selenium chemistry for spatio-selective peptide and protein functionalization.Nat Rev Chem. 2024 Mar;8(3):211-229. doi: 10.1038/s41570-024-00579-1. Epub 2024 Feb 22. Nat Rev Chem. 2024. PMID: 38388838 Review.
References
-
- Carlisle AE, Lee N, Matthew-Onabanjo AN, Spears ME, Park SJ, Youkana D, Doshi MB, Peppers A, Li R, Joseph AB, Smith M, Simin K, Zhu LJ, Greer PL, Shaw LM, Kim D. Selenium detoxification is required for cancer-cell survival. Nature Metabolism. 2020;2:603–611. doi: 10.1038/s42255-020-0224-7. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

