A comparison of 25 complete chloroplast genomes between sister mangrove species Kandelia obovata and Kandelia candel geographically separated by the South China Sea
- PMID: 36684775
- PMCID: PMC9845719
- DOI: 10.3389/fpls.2022.1075353
A comparison of 25 complete chloroplast genomes between sister mangrove species Kandelia obovata and Kandelia candel geographically separated by the South China Sea
Abstract
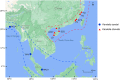
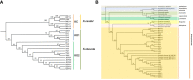
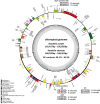
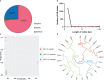
In 2003, Kandelia obovata was identified as a new mangrove species differentiated from Kandelia candel. However, little is known about their chloroplast (cp) genome differences and their possible ecological significance. In this study, 25 whole cp genomes, with seven samples of K. candel from Malaysia, Thailand, and Bangladesh and 18 samples of K. obovata from China, were sequenced for comparison. The cp genomes of both species encoded 128 genes, namely 83 protein-coding genes, 37 tRNA genes, and eight rRNA genes, but the cp genome size of K. obovata was ~2 kb larger than that of K. candle due to the presence of more and longer repeat sequences. Of these, tandem repeats and simple sequence repeats exhibited great differences. Principal component analysis based on indels, and phylogenetic tree analyses constructed with homologous protein genes from the single-copy genes, as well as 38 homologous pair genes among 13 mangrove species, gave strong support to the separation of the two species within the Kandelia genus. Homologous genes ndhD and atpA showed intraspecific consistency and interspecific differences. Molecular dynamics simulations of their corresponding proteins, NAD(P)H dehydrogenase chain 4 (NDH-D) and ATP synthase subunit alpha (ATP-A), predicted them to be significantly different in the functions of photosynthetic electron transport and ATP generation in the two species. These results suggest that the energy requirement was a pivotal factor in their adaptation to differential environments geographically separated by the South China Sea. Our results also provide clues for future research on their physiological and molecular adaptation mechanisms to light and temperature.
Keywords: Kandelia; chloroplast genome; environment adaptation; gene diversity; mangrove; protein dynamics simulation.
Copyright © 2023 Xu, Shen, Zhang, Li, Wang, Chen, Chen, Ng, Islam, Punnarak, Zheng and Zhu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
The complete mitochondrial genome of the Kandelia obovata Sheue, H.Y.Liu & J.W.H.Yong (Rhizophoraceae).Mitochondrial DNA B Resour. 2023 Dec 21;8(12):1440-1444. doi: 10.1080/23802359.2023.2294889. eCollection 2023. Mitochondrial DNA B Resour. 2023. PMID: 38173916 Free PMC article.
-
Complete chloroplast genome sequence of the mangrove species Kandelia obovata and comparative analyses with related species.PeerJ. 2019 Sep 20;7:e7713. doi: 10.7717/peerj.7713. eCollection 2019. PeerJ. 2019. PMID: 31579601 Free PMC article.
-
The complete chloroplast genome of a mangrove Kandelia obovata Sheue, Liu & Yong.Mitochondrial DNA B Resour. 2019 Oct 7;4(2):3414-3415. doi: 10.1080/23802359.2019.1674713. Mitochondrial DNA B Resour. 2019. PMID: 33366018 Free PMC article.
-
Genome-Wide Identification, Characterization, and Expression Analysis of the Copper-Containing Amine Oxidase Gene Family in Mangrove Kandelia obovata.Int J Mol Sci. 2023 Dec 9;24(24):17312. doi: 10.3390/ijms242417312. Int J Mol Sci. 2023. PMID: 38139139 Free PMC article. Review.
-
[Advances in molecular biological studies of the mangrove Kandelia obovata Sheue, Liu & Yong].Sheng Wu Gong Cheng Xue Bao. 2017 Feb 25;33(2):196-204. doi: 10.13345/j.cjb.160272. Sheng Wu Gong Cheng Xue Bao. 2017. PMID: 28956376 Review. Chinese.
Cited by
-
Comparative chloroplast genomes of Incarvillea species (Bignoniaceae) unveiled genomic diversity and shed light on phylogenetic relationships.BMC Plant Biol. 2025 Mar 29;25(1):399. doi: 10.1186/s12870-025-06380-6. BMC Plant Biol. 2025. PMID: 40155865 Free PMC article.
-
The complete mitochondrial genome of the Kandelia obovata Sheue, H.Y.Liu & J.W.H.Yong (Rhizophoraceae).Mitochondrial DNA B Resour. 2023 Dec 21;8(12):1440-1444. doi: 10.1080/23802359.2023.2294889. eCollection 2023. Mitochondrial DNA B Resour. 2023. PMID: 38173916 Free PMC article.
-
Molecular Structure and Variation Characteristics of the Plastomes from Six Malus baccata (L.) Borkh. Individuals and Comparative Genomic Analysis with Other Malus Species.Biomolecules. 2023 Jun 8;13(6):962. doi: 10.3390/biom13060962. Biomolecules. 2023. PMID: 37371542 Free PMC article.
-
Twelve newly assembled jasmine chloroplast genomes: unveiling genomic diversity, phylogenetic relationships and evolutionary patterns among Oleaceae and Jasminum species.BMC Plant Biol. 2024 Apr 25;24(1):331. doi: 10.1186/s12870-024-04995-9. BMC Plant Biol. 2024. PMID: 38664619 Free PMC article.
-
Solanum aculeatissimum and Solanum torvum chloroplast genome sequences: a comparative analysis with other Solanum chloroplast genomes.BMC Genomics. 2024 Apr 26;25(1):412. doi: 10.1186/s12864-024-10190-9. BMC Genomics. 2024. PMID: 38671394 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

