The Delta variant wave in Tunisia: Genetic diversity, spatio-temporal distribution and evidence of the spread of a divergent AY.122 sub-lineage
- PMID: 36684874
- PMCID: PMC9846204
- DOI: 10.3389/fpubh.2022.990832
The Delta variant wave in Tunisia: Genetic diversity, spatio-temporal distribution and evidence of the spread of a divergent AY.122 sub-lineage
Abstract
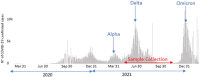
Introduction: The Delta variant posed an increased risk to global public health and rapidly replaced the pre-existent variants worldwide. In this study, the genetic diversity and the spatio-temporal dynamics of 662 SARS-CoV2 genomes obtained during the Delta wave across Tunisia were investigated.
Methods: Viral whole genome and partial S-segment sequencing was performed using Illumina and Sanger platforms, respectively and lineage assignemnt was assessed using Pangolin version 1.2.4 and scorpio version 3.4.X. Phylogenetic and phylogeographic analyses were achieved using IQ-Tree and Beast programs.
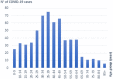
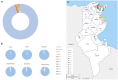
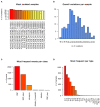
Results: The age distribution of the infected cases showed a large peak between 25 to 50 years. Twelve Delta sub-lineages were detected nation-wide with AY.122 being the predominant variant representing 94.6% of sequences. AY.122 sequences were highly related and shared the amino-acid change ORF1a:A498V, the synonymous mutations 2746T>C, 3037C>T, 8986C>T, 11332A>G in ORF1a and 23683C>T in the S gene with respect to the Wuhan reference genome (NC_045512.2). Spatio-temporal analysis indicates that the larger cities of Nabeul, Tunis and Kairouan constituted epicenters for the AY.122 sub-lineage and subsequent dispersion to the rest of the country.
Discussion: This study adds more knowledge about the Delta variant and sub-variants distribution worldwide by documenting genomic and epidemiological data from Tunisia, a North African region. Such results may be helpful to the understanding of future COVID-19 waves and variants.
Keywords: AY.122; Delta variant; SARS-CoV2; next-generation sequencing; phylogeny; spatio-temporal dynamic.
Copyright © 2023 Haddad-Boubaker, Arbi, Souiai, Chouikha, Fares, Edington, Sims, Camma, Lorusso, Diagne, Diallo, Boubaker, Ferjani, Mastouri, Mhalla, Karray, Gargouri, Bahri, Trabelsi, Kallala, Hannachi, Chaabouni, Smaoui, Meftah, Bouhalila, Foughali, Zribi, Lamari, Touzi, Safer, Alaya, Kahla, Gdoura and Triki.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- World Health Organization. Tracking SARS-CoV2 variants. (2022). Available online at: https://www.who.int/activities/tracking-SARS-CoV-2-variants (accessed February 28, 2022).
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

