Understanding the Spike Protein in COVID-19 Vaccine in Recombinant Vesicular Stomatitis Virus (rVSV) Using Automated Capillary Western Blots
- PMID: 36685032
- PMCID: PMC9843631
- DOI: 10.1021/acsomega.2c06937
Understanding the Spike Protein in COVID-19 Vaccine in Recombinant Vesicular Stomatitis Virus (rVSV) Using Automated Capillary Western Blots
Abstract
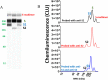
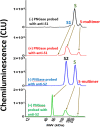
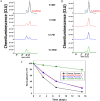
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the viral agent that is responsible for the coronavirus disease-2019 (COVID-19) pandemic. One of the live virus vaccine candidates Merck and Co., Inc. was developing to help combat the pandemic was V590. V590 was a live-attenuated, replication-competent, recombinant vesicular stomatitis virus (rVSV) in which the envelope VSV glycoprotein (G protein) gene was replaced with the gene for the SARS-CoV-2 spike protein (S protein), the protein responsible for viral binding and fusion to the cell membrane. To assist with product and process development, a quantitative Simple Western (SW) assay was successfully developed and phase-appropriately qualified to quantitate the concentration of S protein expressed in V590 samples. A strong correlation was established between potency and S-protein concentration, which suggested that the S-protein SW assay could be used as a proxy for virus productivity optimization with faster data turnaround time (3 h vs 3 days). In addition, unlike potency, the SW assay was able to provide a qualitative profile assessment of the forms of S protein (S protein, S1 subunit, and S multimer) to ensure appropriate levels of S protein were maintained throughout process and product development. Finally, V590 stressed stability studies suggested that time and temperature contributed to the instability of S protein demonstrated by cleavage into its subunits, S1 and S2, and aggregation into S multimer. Both of which could potentially have a deleterious effect on the vaccine immunogenicity.
© 2023 Merck & Co., Inc., Rahway, NJ, USA and its affiliates. Published by American Chemical Society.
Conflict of interest statement
The authors declare the following competing financial interest(s): The authors declare no competing non-financial interests but the following competing financial interests: All authors are/were employees of Merck Sharp & Dohme LLC, a subsidiary of Merck & Co., Inc., Rahway, NJ, USA and may potentially own stock and/or hold stock options in Merck & Co., Inc., Rahway, NJ, USA.
Figures



Similar articles
-
BSL2-compliant lethal mouse model of SARS-CoV-2 and variants of concern to evaluate therapeutics targeting the Spike protein.Front Immunol. 2022 Jul 28;13:919815. doi: 10.3389/fimmu.2022.919815. eCollection 2022. Front Immunol. 2022. PMID: 35967447 Free PMC article.
-
SARS vaccine based on a replication-defective recombinant vesicular stomatitis virus is more potent than one based on a replication-competent vector.Virology. 2008 Jun 20;376(1):165-72. doi: 10.1016/j.virol.2008.03.002. Epub 2008 Apr 8. Virology. 2008. PMID: 18396306 Free PMC article.
-
Preclinical immunogenicity and efficacy of a candidate COVID-19 vaccine based on a vesicular stomatitis virus-SARS-CoV-2 chimera.EBioMedicine. 2022 Aug;82:104203. doi: 10.1016/j.ebiom.2022.104203. Epub 2022 Jul 30. EBioMedicine. 2022. PMID: 35915046 Free PMC article.
-
Severe acute respiratory syndrome vaccine development: experiences of vaccination against avian infectious bronchitis coronavirus.Avian Pathol. 2003 Dec;32(6):567-82. doi: 10.1080/03079450310001621198. Avian Pathol. 2003. PMID: 14676007 Free PMC article. Review.
-
Emergence, evolution, and vaccine production approaches of SARS-CoV-2 virus: Benefits of getting vaccinated and common questions.Saudi J Biol Sci. 2022 Apr;29(4):1981-1997. doi: 10.1016/j.sjbs.2021.12.020. Epub 2021 Dec 13. Saudi J Biol Sci. 2022. PMID: 34924802 Free PMC article. Review.
Cited by
-
Challenges and Opportunities in the Process Development of Chimeric Vaccines.Vaccines (Basel). 2023 Dec 8;11(12):1828. doi: 10.3390/vaccines11121828. Vaccines (Basel). 2023. PMID: 38140232 Free PMC article. Review.
-
Automated, Quantitative Capillary Western Blots to Analyze Host Cell Proteins in COVID-19 Vaccine Produced in Vero Cell Line.Vaccines (Basel). 2024 Dec 5;12(12):1373. doi: 10.3390/vaccines12121373. Vaccines (Basel). 2024. PMID: 39772035 Free PMC article.
-
Functionality and translation fidelity characterization of mRNA vaccines using platform based mass spectrometry detection.NPJ Vaccines. 2025 Feb 23;10(1):38. doi: 10.1038/s41541-025-01082-4. NPJ Vaccines. 2025. PMID: 39988579 Free PMC article.
References
-
- WHO Coronavirus (COVID-19) Dashboard. https://covid19.who.int/ (accessed 04 March, 2022).
-
- Lu R.; Zhao X.; Li J.; Niu P.; Yang B.; Wu H.; Wang W.; Song H.; Huang B.; Zhu N.; Bi Y.; Ma X.; Zhan F.; Wang L.; Hu T.; Zhou H.; Hu Z.; Zhou W.; Zhao L.; Chen J.; Meng Y.; Wang J.; Lin Y.; Yuan J.; Xie Z.; Ma J.; Liu W. J.; Wang D.; Xu W.; Holmes E. C.; Gao G. F.; Wu G.; Chen W.; Shi W.; Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. 10.1016/S0140-6736(20)30251-8. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

