A subnetwork-based framework for prioritizing and evaluating prognostic gene modules from cancer transcriptome data
- PMID: 36685033
- PMCID: PMC9845797
- DOI: 10.1016/j.isci.2022.105915
A subnetwork-based framework for prioritizing and evaluating prognostic gene modules from cancer transcriptome data
Abstract
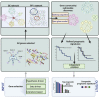
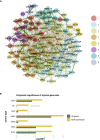
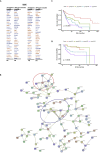
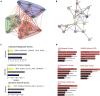
Cancer prognosis prediction is critical to the clinical decision-making process. Currently, the high availability of transcriptome datasets allows us to extract the gene modules with promising prognostic values. However, the biomarker identification is greatly challenged by tumor and patient heterogeneity. In this study, a framework of three subnetwork-based strategies is presented, incorporating hypothesis-driven, data-driven, and literature-based methods with informative visualization to prioritize candidate genes. By applying the proposed approaches to a head and neck squamous cell cancer (HNSCC) transcriptome dataset, we successfully identified multiple HNSCC-specific gene modules with improved prognostic values and mechanism information compared with the standard gene panel selection methods. The proposed framework is general and can be applied to any type of omics data. Overall, the study demonstrates and supports the use of the subnetwork-based approach for distilling reliable and biologically meaningful prognostic factors.
Keywords: Bioinformatics; Cancer; Gene network.
© 2022 The Authors.
Conflict of interest statement
C.H.C. has received honoraria from Sanofi, Merck, and Brooklyn ImmunoTherapeutics, and Exelixis for serving in ad hoc scientific advisory boards. All other authors declare no conflict interest.
Figures

References
-
- Kuksin M., Morel D., Aglave M., Danlos F.X., Marabelle A., Zinovyev A., Gautheret D., Verlingue L. Applications of single-cell and bulk RNA sequencing in onco-immunology. Eur. J. Cancer. 2021;149:193–210. - PubMed
-
- Li K., Wang X., Kuan P.F. Mixture network regularized generalized linear model with feature selection. bioRxiv. 2019:678029. doi: 10.1101/678029. Preprint at. - DOI
LinkOut - more resources
Full Text Sources

