Streptococcus pneumoniae promotes lung cancer development and progression
- PMID: 36685035
- PMCID: PMC9852931
- DOI: 10.1016/j.isci.2022.105923
Streptococcus pneumoniae promotes lung cancer development and progression
Abstract
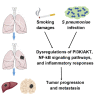
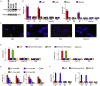
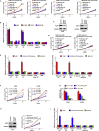
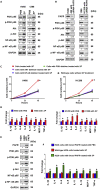
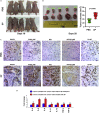
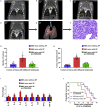
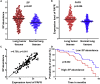
Streptococcus pneumoniae (SP) is associated with lung cancer, yet its role in the tumorigenesis remains uncertain. Herein we find that SP attaches to lung cancer cells via binding pneumococcal surface protein C (PspC) to platelet-activating factor receptor (PAFR). Interaction between PspC and PAFR stimulates cell proliferation and activates PI3K/AKT and nuclear factor kB (NF-kB) signaling pathways, which trigger a pro-inflammatory response. Lung cancer cells infected with SP form larger tumors in BALB/C mice compared to untreated cells. Mice treated with tobacco carcinogen and SP develop more lung tumors and had shorter survival period than mice treated with the carcinogen alone. Mutating PspC or PAFR abolishes tumor-promoting effects of SP. Overabundance of SP is associated with the survival. SP may play a driving role in lung tumorigenesis by activating PI3K/AKT and NF-kB pathways via binding PspC to PAFR and provide a microbial target for diagnosis and treatment of the disease.
Keywords: Bacteriology; Cancer systems biology; Cell; Cell biology.
© 2023 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Mao Q., Jiang F., Yin R., Wang J., Xia W., Dong G., Ma W., Yang Y., Xu L., Hu J. Interplay between the lung microbiome and lung cancer. Cancer Lett. 2018;415:40–48. - PubMed
-
- Hogan D.A., Willger S.D., Dolben E.L., Hampton T.H., Stanton B.A., Morrison H.G., Sogin M.L., Czum J., Ashare A. Analysis of lung microbiota in bronchoalveolar lavage, protected brush and sputum samples from subjects with mild-to-moderate cystic fibrosis lung disease. PLoS One. 2016;11:e0149998. - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

