Comprehensive analysis of cuproptosis-related lncRNAs for prognostic significance and immune microenvironment characterization in hepatocellular carcinoma
- PMID: 36685508
- PMCID: PMC9846072
- DOI: 10.3389/fimmu.2022.991604
Comprehensive analysis of cuproptosis-related lncRNAs for prognostic significance and immune microenvironment characterization in hepatocellular carcinoma
Abstract
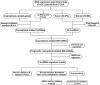
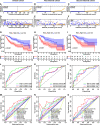
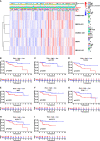
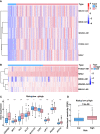
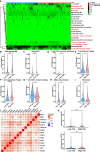
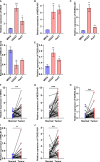
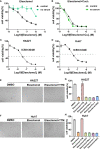
Cuproptosis was characterized as a novel type of programmed cell death. Recently, however, the role of cuproptosis-related long noncoding RNAs (CRLs) in tumors has not yet been studied. Identifying a predictive CRL signature in hepatocellular carcinoma (HCC) and investigating its putative molecular function were the goals of this work. Initially, Pearson's test was used to assess the relationship between lncRNAs and cuproptosis-associated genes obtained from HCC data of The Cancer Genome Atlas (TCGA). By implementing differential expression and univariate Cox analysis, 61 prognostic CRLs were subsequent to the least absolute shrinkage and selection operator (LASSO) Cox regression analysis. A prognostic risk score model was then constructed to evaluate its ability to predict patients' survival when combined with clinicopathological parameters in HCC. The five-lncRNA prognostic signature categorized the HCC patients into high- and low-risk groups. The low-risk group exhibited more sensitivity to elesclomol than the high-risk one. Surprisingly, distinct mitochondrial metabolism pathways connected to cuproptosis and pivotal immune-related pathways were observed between the two groups via gene set enrichment analysis (GSEA). Meanwhile, there were substantial differences between the high-risk group and the low-risk group in terms of tumor-infiltrating immune cells (TIICs). Furthermore, a positive relationship was shown between the risk score and the expression of immune checkpoints. Additionally, differential expression of the five lncRNAs was confirmed in our own HCC samples and cell lines via RT-qPCR. Finally, in vitro assays confirmed that WARS2-AS1 and MKLN1-AS knockdown could sensitize HCC cells to elesclomol-induced cuproptosis. Overall, our predictive signature may predict the prognosis of HCC patients in an independent manner, give a better understanding of how CRLs work in HCC, and offer therapeutic reference for patients with HCC.
Keywords: cuproptosis; hepatocellular carcinoma; immune microenvironment; lncRNA; survival analysis.
Copyright © 2023 Li, Jin, Chen, Zhang, Li, Zhong, Fan and Lin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
A Cuproptosis-Related LncRNA Risk Model for Predicting Prognosis and Immunotherapeutic Efficacy in Patients with Hepatocellular Carcinoma.Biochem Genet. 2024 Jun;62(3):2332-2351. doi: 10.1007/s10528-023-10539-x. Epub 2023 Oct 29. Biochem Genet. 2024. PMID: 37898914
-
A novel cuproptosis-related lncRNA signature predicts the prognosis and immunotherapy for hepatocellular carcinoma.Cancer Biomark. 2023;37(1):13-26. doi: 10.3233/CBM-220259. Cancer Biomark. 2023. PMID: 37005878
-
Comprehensive analysis of cuproptosis-related lncRNAs in immune infiltration and prognosis in hepatocellular carcinoma.BMC Bioinformatics. 2023 Jan 3;24(1):4. doi: 10.1186/s12859-022-05091-1. BMC Bioinformatics. 2023. PMID: 36597032 Free PMC article.
-
Exploring the role of long noncoding RNAs in autophagy and cuproptosis processes via immune pathways in head and neck squamous carcinoma: A systematic review of the literature.Medicine (Baltimore). 2024 Aug 23;103(34):e39335. doi: 10.1097/MD.0000000000039335. Medicine (Baltimore). 2024. PMID: 39183398 Free PMC article.
-
Cuproptosis regulation by long noncoding RNAs: Mechanistic insights and clinical implications in cancer.Arch Biochem Biophys. 2025 Mar;765:110324. doi: 10.1016/j.abb.2025.110324. Epub 2025 Feb 1. Arch Biochem Biophys. 2025. PMID: 39900259 Review.
Cited by
-
Review to Elucidate the Correlation between Cuproptosis-Related Genes and Immune Infiltration for Enhancing the Detection and Treatment of Cervical Cancer.Int J Mol Sci. 2024 Oct 1;25(19):10604. doi: 10.3390/ijms251910604. Int J Mol Sci. 2024. PMID: 39408933 Free PMC article. Review.
-
Prognostic analysis of hepatocellular carcinoma based on cuproptosis -associated lncRNAs.BMC Gastroenterol. 2024 Apr 23;24(1):142. doi: 10.1186/s12876-024-03219-6. BMC Gastroenterol. 2024. PMID: 38654165 Free PMC article.
-
Long non-coding RNA in the regulation of cell death in hepatocellular carcinoma.World J Clin Oncol. 2025 Apr 24;16(4):104061. doi: 10.5306/wjco.v16.i4.104061. World J Clin Oncol. 2025. PMID: 40290684 Free PMC article. Review.
-
Comprehensive Analysis of Disulfidptosis-Related LncRNAs in Molecular Classification, Immune Microenvironment Characterization and Prognosis of Gastric Cancer.Biomedicines. 2023 Nov 28;11(12):3165. doi: 10.3390/biomedicines11123165. Biomedicines. 2023. PMID: 38137387 Free PMC article.
-
The role of mitochondria-related lncRNAs in characterizing the immune landscape and supervising the prognosis of osteosarcoma.J Bone Oncol. 2023 Oct 5;43:100506. doi: 10.1016/j.jbo.2023.100506. eCollection 2023 Dec. J Bone Oncol. 2023. PMID: 37868616 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

