Cc GSDMEa functions the pore-formation in cytomembrane and the regulation on the secretion of IL-lβ in common carp (Cyprinus carpio haematopterus)
- PMID: 36685536
- PMCID: PMC9852915
- DOI: 10.3389/fimmu.2022.1110322
Cc GSDMEa functions the pore-formation in cytomembrane and the regulation on the secretion of IL-lβ in common carp (Cyprinus carpio haematopterus)
Abstract
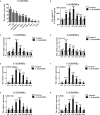
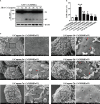
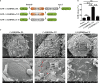
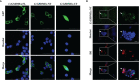
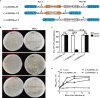
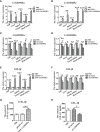
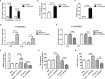
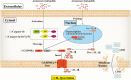
GSDME is the only direct executor of caspase-dependent pyroptosis in both canonical and non-canonical inflammasomes known to date in fish, and plays an important role in anti-bacterial infection and inflammatory response. In order to determine the regulation of GSDMEa on antibacterial infection in innate immune response, the CcGSDMEa gene in common carp (Cyprinus carpio haematopterus) was first identified and characterized, and then its function related to immune defense was investigated. Our results showed that the expressions of CcGSDMEa at the mRNA and protein levels were both significantly increased after Aeromonas hydrophila intraperitoneal infection at the early stage than that in the control group. We found that CcGSDMEa could be cleaved by inflammatory caspase (CcCaspase-1b) and apoptotic caspases (CcCaspase-3a/b and CcCaspase-7a/b). Interestingly, only the CcGSDMEa-NT (1-252 aa) displayed bactericidal activity to Escherichia coli and could punch holes in the membrane of HEK293T cells, whereas CcGSDMEa-FL (1-532 aa) and CcGSDMEa-CT (257-532 aa) showed no above activity and pore-forming ability. Overexpression of CcGSDMEa increased the secretion of CcIL-1β and the release of LDH, and could reduce the A. hydrophila burdens in fish. On the contrary, knockdown of CcGSDMEa reduced the secretion of CcIL-1β and the release of LDH, and could increase the A. hydrophila burdens in fish. Taken together, the elevated expression of CcGSDMEa was a positive immune response to A. hydrophila challenge in fish. CcGSDMEa could perform the pore-formation in cell membrane and the regulation on the secretion of IL-lβ, and further regulate the bacterial clearance in vivo. These results suggested that CcGSDMEa played an important role in immune defense against A. hydrophila and could provide a new insight into understanding the immune mechanism to resist pathogen invasion in teleost.
Keywords: Aeromonas hydrophila; GSDMEa; IL-lβ secretion; caspases; common carp; gasdermin pore.
Copyright © 2023 Zhao, Zhang, Qiao, Gao, Gu, Jiang, Zhu and Kong.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Basu M, Swain B, Maiti NK, Routray P, Samanta M. Inductive expression of toll-like receptor 5 (TLR5) and associated downstream signaling molecules following ligand exposure and bacterial infection in the Indian major carp, mrigal (Cirrhinus mrigala). Fish Shellfish Immunol (2012) 32(1):121–31. doi: 10.1016/j.fsi.2011.10.031 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

