Model-driven experimental design workflow expands understanding of regulatory role of Nac in Escherichia coli
- PMID: 36685725
- PMCID: PMC9853098
- DOI: 10.1093/nargab/lqad006
Model-driven experimental design workflow expands understanding of regulatory role of Nac in Escherichia coli
Abstract
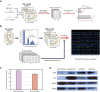
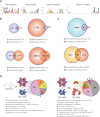
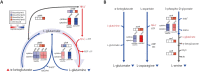
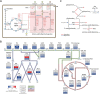
The establishment of experimental conditions for transcriptional regulator network (TRN) reconstruction in bacteria continues to be impeded by the limited knowledge of activating conditions for transcription factors (TFs). Here, we present a novel genome-scale model-driven workflow for designing experimental conditions, which optimally activate specific TFs. Our model-driven workflow was applied to elucidate transcriptional regulation under nitrogen limitation by Nac and NtrC, in Escherichia coli. We comprehensively predict alternative nitrogen sources, including cytosine and cytidine, which trigger differential activation of Nac using a model-driven workflow. In accordance with the prediction, genome-wide measurements with ChIP-exo and RNA-seq were performed. Integrative data analysis reveals that the Nac and NtrC regulons consist of 97 and 43 genes under alternative nitrogen conditions, respectively. Functional analysis of Nac at the transcriptional level showed that Nac directly down-regulates amino acid biosynthesis and restores expression of tricarboxylic acid (TCA) cycle genes to alleviate nitrogen-limiting stress. We also demonstrate that both TFs coherently modulate α-ketoglutarate accumulation stress due to nitrogen limitation by co-activating amino acid and diamine degradation pathways. A systems-biology approach provided a detailed and quantitative understanding of both TF's roles and how nitrogen and carbon metabolic networks respond complementarily to nitrogen-limiting stress.
© The Author(s) 2023. Published by Oxford University Press on behalf of NAR Genomics and Bioinformatics.
Figures

Similar articles
-
Systematic discovery of uncharacterized transcription factors in Escherichia coli K-12 MG1655.Nucleic Acids Res. 2018 Nov 16;46(20):10682-10696. doi: 10.1093/nar/gky752. Nucleic Acids Res. 2018. PMID: 30137486 Free PMC article.
-
Genome-wide Reconstruction of OxyR and SoxRS Transcriptional Regulatory Networks under Oxidative Stress in Escherichia coli K-12 MG1655.Cell Rep. 2015 Aug 25;12(8):1289-99. doi: 10.1016/j.celrep.2015.07.043. Epub 2015 Aug 13. Cell Rep. 2015. PMID: 26279566
-
Elucidation of Regulatory Modes for Five Two-Component Systems in Escherichia coli Reveals Novel Relationships.mSystems. 2020 Nov 10;5(6):e00980-20. doi: 10.1128/mSystems.00980-20. mSystems. 2020. PMID: 33172971 Free PMC article.
-
The interplay between ABA/ethylene and NAC TFs in tomato fruit ripening: a review.Plant Mol Biol. 2021 Jun;106(3):223-238. doi: 10.1007/s11103-021-01128-w. Epub 2021 Feb 25. Plant Mol Biol. 2021. PMID: 33634368 Review.
-
A NAC for regulating metabolism: the nitrogen assimilation control protein (NAC) from Klebsiella pneumoniae.J Bacteriol. 2010 Oct;192(19):4801-11. doi: 10.1128/JB.00266-10. Epub 2010 Jul 30. J Bacteriol. 2010. PMID: 20675498 Free PMC article. Review.
Cited by
-
Unveiling the novel regulatory roles of RpoD-family sigma factors in Salmonella Typhimurium heat shock response through systems biology approaches.PLoS Genet. 2024 Oct 29;20(10):e1011464. doi: 10.1371/journal.pgen.1011464. eCollection 2024 Oct. PLoS Genet. 2024. PMID: 39471211 Free PMC article.
-
ChIP-mini: a low-input ChIP-exo protocol for elucidating DNA-binding protein dynamics in intracellular pathogens.Nucleic Acids Res. 2025 Jan 24;53(3):gkaf009. doi: 10.1093/nar/gkaf009. Nucleic Acids Res. 2025. PMID: 39868540 Free PMC article.
-
Revealing systematic changes in the transcriptome during the transition from exponential growth to stationary phase.mSystems. 2025 Jan 21;10(1):e0131524. doi: 10.1128/msystems.01315-24. Epub 2024 Dec 23. mSystems. 2025. PMID: 39714213 Free PMC article.
References
-
- Cho B.K., Knight E.M., Palsson B.O.. Transcriptional regulation of the fad regulon genes of Escherichia coli by ArcA. Microbiology. 2006; 152:2207–2219. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

