Identification and experimental validation of key m6A modification regulators as potential biomarkers of osteoporosis
- PMID: 36685841
- PMCID: PMC9852729
- DOI: 10.3389/fgene.2022.1072948
Identification and experimental validation of key m6A modification regulators as potential biomarkers of osteoporosis
Abstract
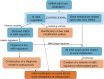
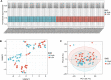
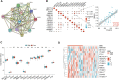
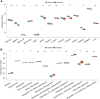
Osteoporosis (OP) is a severe systemic bone metabolic disease that occurs worldwide. During the coronavirus pandemic, prioritization of urgent services and delay of elective care attenuated routine screening and monitoring of OP patients. There is an urgent need for novel and effective screening diagnostic biomarkers that require minimal technical and time investments. Several studies have indicated that N6-methyladenosine (m6A) regulators play essential roles in metabolic diseases, including OP. The aim of this study was to identify key m6A regulators as biomarkers of OP through gene expression data analysis and experimental verification. GSE56815 dataset was served as the training dataset for 40 women with high bone mineral density (BMD) and 40 women with low BMD. The expression levels of 14 major m6A regulators were analyzed to screen for differentially expressed m6A regulators in the two groups. The impact of m6A modification on bone metabolism microenvironment characteristics was explored, including osteoblast-related and osteoclast-related gene sets. Most m6A regulators and bone metabolism-related gene sets were dysregulated in the low-BMD samples, and their relationship was also tightly linked. In addition, consensus cluster analysis was performed, and two distinct m6A modification patterns were identified in the low-BMD samples. Subsequently, by univariate and multivariate logistic regression analyses, we identified four key m6A regulators, namely, METTL16, CBLL1, FTO, and YTHDF2. We built a diagnostic model based on the four m6A regulators. CBLL1 and YTHDF2 were protective factors, whereas METTL16 and FTO were risk factors, and the ROC curve and test dataset validated that this model had moderate accuracy in distinguishing high- and low-BMD samples. Furthermore, a regulatory network was constructed of the four hub m6A regulators and 26 m6A target bone metabolism-related genes, which enhanced our understanding of the regulatory mechanisms of m6A modification in OP. Finally, the expression of the four key m6A regulators was validated in vivo and in vitro, which is consistent with the bioinformatic analysis results. Our findings identified four key m6A regulators that are essential for bone metabolism and have specific diagnostic value in OP. These modules could be used as biomarkers of OP in the future.
Keywords: M6A; RNA modification; biomarker; bone metabolism; osteoclast; osteoporosis.
Copyright © 2023 Qiao, Li, Liu, Zhang, Liu and Zheng.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Unraveling the Role of N6-Methylation Modification: From Bone Biology to Osteoporosis.Int J Med Sci. 2025 May 8;22(11):2545-2559. doi: 10.7150/ijms.108763. eCollection 2025. Int J Med Sci. 2025. PMID: 40520903 Free PMC article. Review.
-
Comprehensive analysis of the m6A-related molecular patterns and diagnostic biomarkers in osteoporosis.Front Endocrinol (Lausanne). 2022 Aug 10;13:957742. doi: 10.3389/fendo.2022.957742. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 36034449 Free PMC article.
-
Characteristics of n6-methyladenosine (m6A) regulators and role of FTO/TNC in scleroderma.Gene. 2024 Feb 5;894:147989. doi: 10.1016/j.gene.2023.147989. Epub 2023 Nov 14. Gene. 2024. PMID: 37972699
-
Landscape analysis of m6A modification regulators related biological functions and immune characteristics in myasthenia gravis.J Transl Med. 2023 Mar 2;21(1):166. doi: 10.1186/s12967-023-03947-5. J Transl Med. 2023. PMID: 36864526 Free PMC article.
-
Research advances of N6-methyladenosine in diagnosis and therapy of pancreatic cancer.J Clin Lab Anal. 2022 Sep;36(9):e24611. doi: 10.1002/jcla.24611. Epub 2022 Jul 15. J Clin Lab Anal. 2022. PMID: 35837987 Free PMC article. Review.
Cited by
-
FTO-associated osteoclastogenesis promotes alveolar bone resorption in apical periodontitis male rat via the HK1/USP14/RANK pathway.Nat Commun. 2025 Feb 11;16(1):1519. doi: 10.1038/s41467-025-56615-1. Nat Commun. 2025. PMID: 39934129 Free PMC article.
-
Epigenetic Mechanisms in Osteoporosis: Exploring the Power of m6A RNA Modification.J Cell Mol Med. 2025 Jan;29(1):e70344. doi: 10.1111/jcmm.70344. J Cell Mol Med. 2025. PMID: 39779466 Free PMC article. Review.
-
Unraveling the Role of N6-Methylation Modification: From Bone Biology to Osteoporosis.Int J Med Sci. 2025 May 8;22(11):2545-2559. doi: 10.7150/ijms.108763. eCollection 2025. Int J Med Sci. 2025. PMID: 40520903 Free PMC article. Review.
-
A Machine Learning Framework for Screening Plasma Cell-Associated Feature Genes to Estimate Osteoporosis Risk and Treatment Vulnerability.Biochem Genet. 2025 Aug;63(4):3117-3138. doi: 10.1007/s10528-024-10861-y. Epub 2024 Jun 19. Biochem Genet. 2025. PMID: 38898268
-
Recent advances of m6A methylation in skeletal system disease.J Transl Med. 2024 Feb 14;22(1):153. doi: 10.1186/s12967-024-04944-y. J Transl Med. 2024. PMID: 38355483 Free PMC article. Review.
References
-
- Camacho P. M., Petak S. M., Binkley N., Diab D. L., Eldeiry L. S., Farooki A., et al. (2020). American association of clinical endocrinologists/American college of endocrinology clinical practice guidelines for the diagnosis and treatment of postmenopausal osteoporosis-2020 update. Endocr. Pract. 26, 1–46. 10.4158/GL-2020-0524SUPPL - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

