Bioinformatic analysis and machine learning to identify the diagnostic biomarkers and immune infiltration in adenomyosis
- PMID: 36685847
- PMCID: PMC9845720
- DOI: 10.3389/fgene.2022.1082709
Bioinformatic analysis and machine learning to identify the diagnostic biomarkers and immune infiltration in adenomyosis
Abstract
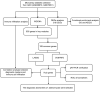
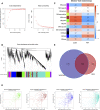
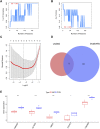
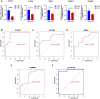
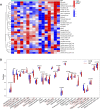
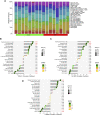
Background: Adenomyosis is a hormone-dependent benign gynecological disease characterized by the invasion of the endometrium into the myometrium. Women with adenomyosis can suffer from abnormal uterine bleeding, severe pelvic pain, and subfertility or infertility, which can interfere with their quality of life. However, effective diagnostic biomarkers for adenomyosis are currently lacking. The aim of this study is to explore the mechanism of adenomyosis by identifying biomarkers and potential therapeutic targets for adenomyosis and analyzing their correlation with immune infiltration in adenomyosis. Methods: Two datasets, GSE78851 and GSE68870, were downloaded and merged for differential expression analysis and functional enrichment analysis using R software. Weighted gene co-expression network analysis (WGCNA), the least absolute shrinkage and selection operator (LASSO), and support vector machine-recursive feature elimination (SVE-RFE) were combined to explore candidate genes. Quantitative reverse transcriptase PCR (qRT-PCR) was conducted to verify the biomarkers and receiver operating characteristic curve analysis was used to assess the diagnostic value of each biomarker. Single-sample Gene Set Enrichment Analysis (ssGSEA) and CIBERSORT were used to explore immune cell infiltration in adenomyosis and the correlation between diagnostic biomarkers and immune cells. Results: A total of 318 genes were differentially expressed. Through the analysis of differentially expressed genes and WGCNA, we obtained 189 adenomyosis-related genes. After utilizing the LASSO and SVM-RFE algorithms, four hub genes, namely, six-transmembrane epithelial antigen of the prostate-1 (STEAP1), translocase of outer mitochondrial membrane 20 (TOMM20), glycosyltransferase eight domain-containing 2 (GLT8D2), and NME/NM23 family member 5 (NME5) expressed in nucleoside-diphosphate kinase, were identified and verified by qRT-PCR. Immune infiltration analysis indicated that T helper 17 cells, CD56dim natural killer cells, monocytes, and memory B-cell may be associated with the occurrence of adenomyosis. There were significant correlations between the diagnostic biomarkers and immune cells. Conclusion: STEAP1, TOMM20, GLT8D2, and NME5 were identified as potential biomarkers and therapeutic targets for adenomyosis. Immune infiltration may contribute to the onset and progression of adenomyosis.
Keywords: WGCNA; adenomyosis; bioinformatics analysis; diagnostic markers; immune infiltration; machine learning.
Copyright © 2023 Liu, Yin, Guan and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources
Research Materials

