Whole-genome resequencing reveals genetic diversity and selection characteristics of dairy goat
- PMID: 36685859
- PMCID: PMC9852865
- DOI: 10.3389/fgene.2022.1044017
Whole-genome resequencing reveals genetic diversity and selection characteristics of dairy goat
Abstract
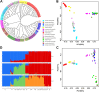
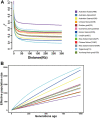
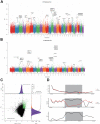
The dairy goat is one of the earliest dairy livestock species, which plays an important role in the economic development, especially for developing countries. With the development of agricultural civilization, dairy goats have been widely distributed across the world. However, few studies have been conducted on the specific characteristics of dairy goat. In this study, we collected the whole-genome data of 89 goat individuals by sequencing 48 goats and employing 41 publicly available goats, including five dairy goat breeds (Saanen, Nubian, Alpine, Toggenburg, and Guanzhong dairy goat; n = 24, 15, 11, 6, 6), and three goat breeds (Guishan goat, Longlin goat, Yunshang Black goat; n = 6, 15, 6). Through compared the genomes of dairy goat and non-dairy goat to analyze genetic diversity and selection characteristics of dairy goat. The results show that the eight goats could be divided into three subgroups of European, African, and Chinese indigenous goat populations, and we also found that Australian Nubian, Toggenburg, and Australian Alpine had the highest linkage disequilibrium, the lowest level of nucleotide diversity, and a higher inbreeding coefficient, indicating that they were strongly artificially selected. In addition, we identified several candidate genes related to the specificity of dairy goat, particularly genes associated with milk production traits (GHR, DGAT2, ELF5, GLYCAM1, ACSBG2, ACSS2), reproduction traits (TSHR, TSHB, PTGS2, ESR2), immunity traits (JAK1, POU2F2, LRRC66). Our results provide not only insights into the evolutionary history and breed characteristics of dairy goat, but also valuable information for the implementation and improvement of dairy goat cross breeding program.
Keywords: dairy goat; genetic diversity; milk production traits; population structure; selective signal.
Copyright © 2023 Xiong, Bao, Hu, Shang and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Whole-genome resequencing reveals diversity and selective signals in Longlin goat.Gene. 2021 Mar 1;771:145371. doi: 10.1016/j.gene.2020.145371. Epub 2020 Dec 17. Gene. 2021. PMID: 33346103
-
Whole-genome resequencing of native and imported dairy goat identifies genes associated with productivity and immunity.Front Vet Sci. 2024 Jul 8;11:1409282. doi: 10.3389/fvets.2024.1409282. eCollection 2024. Front Vet Sci. 2024. PMID: 39040818 Free PMC article.
-
Whole-genome resequencing reveals candidate genes associated with milk production trait in Guanzhong dairy goats.Anim Genet. 2024 Feb;55(1):168-172. doi: 10.1111/age.13380. Epub 2023 Dec 13. Anim Genet. 2024. PMID: 38093616
-
Status and prospects of the dairy goat industry in the United States.J Anim Sci. 1996 May;74(5):1173-81. doi: 10.2527/1996.7451173x. J Anim Sci. 1996. PMID: 8726751 Review.
-
Genomic advancements in goat breeding: enhancing productivity, disease resistance, and sustainability in India's rural economy.Mamm Genome. 2025 May 28. doi: 10.1007/s00335-025-10138-8. Online ahead of print. Mamm Genome. 2025. PMID: 40434651 Review.
Cited by
-
Examination of homozygosity runs and selection signatures in native goat breeds of Henan, China.BMC Genomics. 2024 Dec 7;25(1):1184. doi: 10.1186/s12864-024-11098-0. BMC Genomics. 2024. PMID: 39643897 Free PMC article.
-
Genetic diversity and reproductive trait selection signal analysis of Bohuai goats based on whole genome sequencing.BMC Genomics. 2025 Jul 26;26(1):693. doi: 10.1186/s12864-025-11878-2. BMC Genomics. 2025. PMID: 40713489 Free PMC article.
-
Genome-Wide Runs of Homozygosity Reveal Inbreeding Levels and Trait-Associated Candidate Genes in Diverse Sheep Breeds.Genes (Basel). 2025 Mar 7;16(3):316. doi: 10.3390/genes16030316. Genes (Basel). 2025. PMID: 40149467 Free PMC article.
-
Genetic Diversity, Population Structure and Selection Signature in Begait Goats Revealed by Whole-Genome Sequencing.Animals (Basel). 2024 Jan 18;14(2):307. doi: 10.3390/ani14020307. Animals (Basel). 2024. PMID: 38254476 Free PMC article.
-
Examination of Runs of Homozygosity Distribution Patterns and Relevant Candidate Genes of Potential Economic Interest in Russian Goat Breeds Using Whole-Genome Sequencing.Genes (Basel). 2025 May 24;16(6):631. doi: 10.3390/genes16060631. Genes (Basel). 2025. PMID: 40565522 Free PMC article.
References
-
- Adams H. A., Sonstegard T. S., VanRaden P. M., Null D. J., Van Tassell C. P., Larkin D. M., et al. (2016). Identification of a nonsense mutation in APAF1 that is likely causal for a decrease in reproductive efficiency in Holstein dairy cattle. J. Dairy Sci. 99 (8), 6693–6701. 10.3168/jds.2015-10517 - DOI - PubMed
-
- An X. P., Song S. G., Hou J. X., Zhu C. M., Peng J. X., Liu X. Q., et al. (2011). Polymorphism identification in goat DGAT2 gene and association analysis with milk yield and fat percentage. Small Ruminant Res. 100 (2-3), 107–112. 10.1016/j.smallrumres.2011.05.017 - DOI
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

