An integrative analysis of an lncRNA-mRNA competing endogenous RNA network to identify functional lncRNAs in uterine leiomyomas with RNA sequencing
- PMID: 36685910
- PMCID: PMC9845257
- DOI: 10.3389/fgene.2022.1053845
An integrative analysis of an lncRNA-mRNA competing endogenous RNA network to identify functional lncRNAs in uterine leiomyomas with RNA sequencing
Abstract
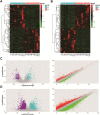
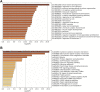
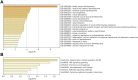
Objective: To explore the functions of mRNAs and lncRNAs in the occurrence of uterine leiomyomas (ULs) and further clarify the pathogenesis of UL by detecting the differential expression of mRNAs and lncRNAs in 10 cases of UL tissues and surrounding normal myometrial tissues by high-throughput RNA sequencing. Methods: The tissue samples of 10 patients who underwent hysterectomy for UL in Lianyungang Maternal and Child Health Hospital from January 2016 to December 2021 were collected. The differentially expressed mRNAs (DEmRNAs) and lncRNAs (DElncRNAs) were identified and further analyzed by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. The protein-protein interaction network (PPI) was constructed in Cytoscape software. Functional annotation of the nearby target cis-DEmRNAs of DElncRNAs was performed with the Database for Annotation, Visualization, and Integrated Discovery (DAVID) (https://david.ncifcrf.gov/). Meanwhile, the co-expression network of DElncRNA-DEmRNA was constructed in Cytoscape software. Results: A total of 553 DElncRNAs (283 upregulated DElncRNAs and 270 downregulated DElncRNAs) and 3,293 DEmRNAs (1,632 upregulated DEmRNAs and 1,661 downregulated DEmRNAs) were obtained. GO pathway enrichment analysis revealed that several important pathways were significantly enriched in UL such as blood vessel development, regulation of ion transport, and external encapsulating structure organization. In addition, cytokine-cytokine receptor interaction, neuroactive ligand-receptor interaction, and complement and coagulation cascades were significantly enriched in KEGG pathway enrichment analysis. A total of 409 DElncRNAs-nearby-targeted DEmRNA pairs were detected, which included 118 DElncRNAs and 136 DEmRNAs. Finally, we found that the top two DElncRNAs with the most nearby DEmRNAs were BISPR and AC012531.1. Conclusion: These results suggested that 3,293 DEmRNAs and 553 DElncRNAs were differentially expressed in UL tissue and normal myometrium tissue, which might be candidate-identified therapeutic and prognostic targets for UL and be considered as offering several possible mechanisms and pathogenesis of UL in the future.
Keywords: high-throughput sequencing; long non-coding RNA; mRNAs; miRNAs; uterine leiomyomas.
Copyright © 2023 Meng, Ji, Chen, Wang and Hua.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources

