Identification of ferroptosis, necroptosis, and pyroptosis-associated genes in periodontitis-affected human periodontal tissue using integrated bioinformatic analysis
- PMID: 36686646
- PMCID: PMC9852864
- DOI: 10.3389/fphar.2022.1098851
Identification of ferroptosis, necroptosis, and pyroptosis-associated genes in periodontitis-affected human periodontal tissue using integrated bioinformatic analysis
Abstract
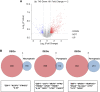
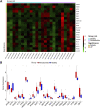
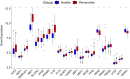
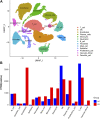
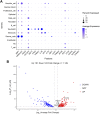
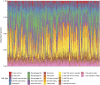
Introduction: Periodontitis is a chronic inflammatory oral disease that destroys soft and hard periodontal support tissues. Multiple cell death modes including apoptosis, necroptosis, pyroptosis, and ferroptosis play a crucial role in the pathogenicity of inflammatory diseases. This study aimed to identify genes associated with ferroptosis, necroptosis, and pyroptosis in different cells present in the periodontium of periodontitis patients. Methods: Gingival tissues' mRNA sequencing dataset GSE173078 of 12 healthy control and 12 periodontitis patients' and the microarray dataset GSE10334 of 63 healthy controls and 64 periodontitis patients' were obtained from Gene Expression Omnibus (GEO) database. A total of 910 differentially expressed genes (DEGs) obtained in GSE173078 were intersected with necroptosis, pyroptosis, and ferroptosis-related genes to obtain the differential genes associated with cell death (DCDEGs), and the expression levels of 21 differential genes associated with cell death were verified with dataset GSE10334. Results: Bioinformatic analysis revealed 21 differential genes associated with cell death attributed to ferroptosis, pyroptosis, and necroptosis in periodontitis patients compared with healthy controls. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses revealed that 21 differential genes associated with cell death were related to various cellular and immunological pathways including inflammatory responses, necroptosis, and osteoclast differentiation. Additionally, the single-cell RNA (scRNA) sequencing data GSE171213 of 4 healthy controls and 5 periodontitis patients' periodontal tissue was analyzed to obtain cell clustering and cell types attributed to differential genes associated with cell death. We found that among 21 DCDEGs, SLC2A3, FPR2, TREM1, and IL1B were mainly upregulated in neutrophils present in the periodontium of periodontitis patients. Gene overlapping analysis revealed that IL-1B is related to necroptosis and pyroptosis, TREM1 and FPR2 are related to pyroptosis, and SLC2A3 is related to ferroptosis. Finally, we utilized the CIBERSORT algorithm to assess the association between DCDEGs and immune infiltration phenotypes, based on the gene expression profile of GSE10334. The results revealed that the upregulated SLC2A3, FPR2, TREM1, and IL1B were positively correlated with neutrophil infiltration in the periodontium. Discussion: The findings provide upregulated SLC2A3, FPR2, TREM1, and IL1B in neutrophils as a future research direction on the mode and mechanism of cell death in periodontitis and their role in disease pathogenicity.
Keywords: bioinformatic analysis; ferroptosis; necroptosis; neutrophils; periodontitis; pyroptosis.
Copyright © 2023 Pan, Li, He, Cheng, Li and Pathak.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Ansari J., Senchenkova E. Y., Vital S. A., Al-Yafeai Z., Kaur G., Sparkenbaugh E. M., et al. (2021). Targeting the AnxA1/Fpr2/ALX pathway regulates neutrophil function, promoting thromboinflammation resolution in sickle cell disease. Blood 137, 1538–1549. 10.1182/blood.2020009166 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

