Upregulation of complement proteins in lung cancer cells mediates tumor progression
- PMID: 36686777
- PMCID: PMC9849673
- DOI: 10.3389/fonc.2022.1045690
Upregulation of complement proteins in lung cancer cells mediates tumor progression
Abstract
Introduction: In vivo, cancer cells respond to signals from the tumor microenvironment resulting in changes in expression of proteins that promote tumor progression and suppress anti-tumor immunity. This study employed an orthotopic immunocompetent model of lung cancer to define pathways that are altered in cancer cells recovered from tumors compared to cells grown in culture.
Methods: Studies used four murine cell lines implanted into the lungs of syngeneic mice. Cancer cells were recovered using FACS, and transcriptional changes compared to cells grown in culture were determined by RNA-seq.
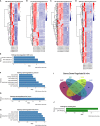
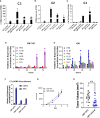
Results: Changes in interferon response, antigen presentation and cytokine signaling were observed in all tumors. In addition, we observed induction of the complement pathway. We previously demonstrated that activation of complement is critical for tumor progression in this model. Complement can play both a pro-tumorigenic role through production of anaphylatoxins, and an anti-tumorigenic role by promoting complement-mediated cell killing of cancer cells. While complement proteins are produced by the liver, expression of complement proteins by cancer cells has been described. Silencing cancer cell-specific C3 inhibited tumor growth In vivo. We hypothesized that induction of complement regulatory proteins was critical for blocking the anti-tumor effects of complement activation. Silencing complement regulatory proteins also inhibited tumor growth, with different regulatory proteins acting in a cell-specific manner.
Discussion: Based on these data we propose that localized induction of complement in cancer cells is a common feature of lung tumors that promotes tumor progression, with induction of complement regulatory proteins protecting cells from complement mediated-cell killing.
Keywords: NSCLC; RNA sequencing; complement; factor H (FH); tumor microenvironment.
Copyright © 2023 Kleczko, Poczobutt, Navarro, Laskowski, Johnson, Korpela, Gurule, Heasley, Hopp, Weiser-Evans, Gottlin, Bushey, Campa, Patz, Thurman and Nemenoff.
Conflict of interest statement
Author EP patent issued to Grid Therapeuticss for inhibitors of complement factor H; chief executive officer, founder, and board member of Grid Therapeutics; stock or stock options in Grid Therapeutics. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Gettinger SN, Horn L, Gandhi L, Spigel DR, Antonia SJ, Rizvi NA, et al. Overall survival and long-term safety of nivolumab (Anti-programmed death 1 antibody, bms-936558, Ono-4538) in patients with previously treated advanced non-Small-Cell lung cancer. J Clin Oncol (2015) 33(18):2004–12. doi: 10.1200/JCO.2014.58.3708 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

