Palindrome-Embedded Hairpin Structure and Its Target-Catalyzed Padlock Cyclization for Label-Free MicroRNA-Initiated Rolling Circle Amplification
- PMID: 36687024
- PMCID: PMC9850459
- DOI: 10.1021/acsomega.2c06532
Palindrome-Embedded Hairpin Structure and Its Target-Catalyzed Padlock Cyclization for Label-Free MicroRNA-Initiated Rolling Circle Amplification
Abstract
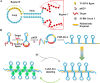
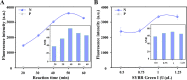
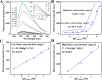
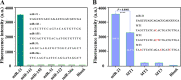
Highly sensitive detection of microRNAs (miRNAs) is of great significance in early diagnosis of cancers. Here, we develop a palindrome-embedded hairpin structure and its target-catalyzed padlock cyclization for rolling circle amplification, named PHP-RCA for simplicity, which can be applied in label-free ultrasensitive detection of miRNA. PHP-RCA is a facile system that consists of only an oligonucleotide probe with a palindrome-embedded hairpin structure (PHP). The two ends of PHP were extended as overhangs and designed with the complementary sequences of the target. Hence, the phosphorylated PHP can be cyclized by T4 DNA ligase in the presence of the target that serves as the ligation template. This ligation has formed a palindrome-embedded dumbbell-shaped probe (PDP) that allows phi29 polymerase to perform a typical target-primed RCA on PDP by taking miRNA as a primer, resulting in the production of a lengthy tandem repeat. Benefits from the palindromic sequences and hairpin-shaped structure in padlock double-stranded structures can be infinitely produced during the RCA reaction and provide numerous binding sites for SYBR Green I, a double-stranded dye, achieving a sharp response signal for label-free target detection. We have demonstrated that the proposed system exhibits a good linear range from 0.1 fM to 5 nM with a low detection limit of 0.1 fM, and the non-target miRNA can be clearly distinguished. The advantages of high efficiency, label-free signaling, and the use of only one oligonucleotide component make the PHP-RCA suitable for ultrasensitive, economic, and convenient detection of target miRNAs. This simple and powerful system is expected to provide a promising platform for tumor diagnosis, prognosis, and therapy.
© 2023 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

Similar articles
-
Target-catalyzed hairpin structure-mediated padlock cyclization for ultrasensitive rolling circle amplification.Talanta. 2019 Nov 1;204:29-35. doi: 10.1016/j.talanta.2019.05.057. Epub 2019 May 15. Talanta. 2019. PMID: 31357296
-
A label-free fluorescent biosensor for amplified detection of T4 polynucleotide kinase activity based on rolling circle amplification and catalytic hairpin assembly.Spectrochim Acta A Mol Biomol Spectrosc. 2023 Jan 15;285:121938. doi: 10.1016/j.saa.2022.121938. Epub 2022 Oct 2. Spectrochim Acta A Mol Biomol Spectrosc. 2023. PMID: 36209712
-
Label-free fluorometric detection of microRNA using isothermal rolling circle amplification generating tandem G-quadruplex.Analyst. 2020 Sep 14;145(18):6130-6137. doi: 10.1039/d0an01329c. Analyst. 2020. PMID: 32869779
-
Recent advances in rolling circle amplification-based biosensing strategies-A review.Anal Chim Acta. 2021 Mar 1;1148:238187. doi: 10.1016/j.aca.2020.12.062. Epub 2020 Dec 31. Anal Chim Acta. 2021. PMID: 33516384 Review.
-
A fluorescence quenching-recovery sensor based on RCA for the specific analysis of Fusobacterium nucleatum. nucleatum.Anal Biochem. 2020 Sep 1;604:113808. doi: 10.1016/j.ab.2020.113808. Epub 2020 Jul 3. Anal Biochem. 2020. PMID: 32622974 Review.
Cited by
-
Target recycling amplification (TRA) combined with multiple strand displacement amplification (SDA) for sensitive detection of Epstein-Barr virus microRNA.Pract Lab Med. 2025 Aug 6;46:e00496. doi: 10.1016/j.plabm.2025.e00496. eCollection 2025 Sep. Pract Lab Med. 2025. PMID: 40823120 Free PMC article.
References
LinkOut - more resources
Full Text Sources

