SARS-CoV-2 variants induce distinct disease and impact in the bone marrow and thymus of mice
- PMID: 36687317
- PMCID: PMC9838028
- DOI: 10.1016/j.isci.2023.105972
SARS-CoV-2 variants induce distinct disease and impact in the bone marrow and thymus of mice
Abstract
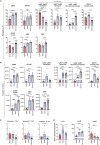
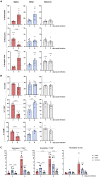
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has evolved to variants associated with milder disease. We employed the k18-hACE2 mouse model to study how differences in the course of infection by SARS-CoV-2 variants alpha, delta, and omicron relate to tissue pathology and the immune response triggered. We documented a variant-specific pattern of infection severity, inducing discrete lung and blood immune responses and differentially impacting primary lymphoid organs. Infections with variants alpha and delta promoted bone marrow (BM) emergency myelopoiesis, with blood and lung neutrophilia. The defects in the BM hematopoietic compartment extended to the thymus, with the infection by the alpha variant provoking a marked thymic atrophy. Importantly, the changes in the immune responses correlated with the severity of infection. Our study provides a comprehensive platform to investigate the modulation of disease by SARS-CoV-2 variants and underscores the impact of this infection on the function of primary lymphoid organs.
Keywords: Components of the immune system; Immunology; Pathology; Virology.
© 2023 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






Similar articles
-
Comparison of the Pathogenicity of SARS-CoV-2 Delta and Omicron Variants by Analyzing the Expression Patterns of Immune Response Genes in K18-hACE2 Transgenic Mice.Front Biosci (Landmark Ed). 2022 Nov 30;27(11):316. doi: 10.31083/j.fbl2711316. Front Biosci (Landmark Ed). 2022. PMID: 36472114
-
SARS-CoV-2 Causes Lung Infection without Severe Disease in Human ACE2 Knock-In Mice.J Virol. 2022 Jan 12;96(1):e0151121. doi: 10.1128/JVI.01511-21. Epub 2021 Oct 20. J Virol. 2022. PMID: 34668780 Free PMC article.
-
The K18-Human ACE2 Transgenic Mouse Model Recapitulates Non-severe and Severe COVID-19 in Response to an Infectious Dose of the SARS-CoV-2 Virus.J Virol. 2022 Jan 12;96(1):e0096421. doi: 10.1128/JVI.00964-21. Epub 2021 Oct 20. J Virol. 2022. PMID: 34668775 Free PMC article.
-
Early pathogenesis profiles across SARS-CoV-2 variants in K18-hACE2 mice revealed differential triggers of lung damages.Front Immunol. 2022 Oct 27;13:950666. doi: 10.3389/fimmu.2022.950666. eCollection 2022. Front Immunol. 2022. PMID: 36389747 Free PMC article.
-
Nasal delivery of broadly neutralizing antibodies protects mice from lethal challenge with SARS-CoV-2 delta and omicron variants.Virol Sin. 2022 Apr;37(2):238-247. doi: 10.1016/j.virs.2022.02.005. Epub 2022 Feb 18. Virol Sin. 2022. PMID: 35527227 Free PMC article.
Cited by
-
In Silico Binding of 2-Aminocyclobutanones to SARS-CoV-2 Nsp13 Helicase and Demonstration of Antiviral Activity.Int J Mol Sci. 2023 Mar 7;24(6):5120. doi: 10.3390/ijms24065120. Int J Mol Sci. 2023. PMID: 36982188 Free PMC article.
-
Protocol for infecting and monitoring susceptible k18-hACE2 mice with SARS-CoV-2.STAR Protoc. 2023 Apr 27;4(2):102303. doi: 10.1016/j.xpro.2023.102303. Online ahead of print. STAR Protoc. 2023. PMID: 37178116 Free PMC article.
-
The impact of COVID-19 on accelerating of immunosenescence and brain aging.Front Cell Neurosci. 2024 Dec 10;18:1471192. doi: 10.3389/fncel.2024.1471192. eCollection 2024. Front Cell Neurosci. 2024. PMID: 39720706 Free PMC article. Review.
-
The impact of SARS-CoV-2 infection on immunity reconstitution among pediatric patients after allogeneic hematopoietic stem cell transplantation: a propensity score-matched analysis.Front Pediatr. 2024 Nov 21;12:1489648. doi: 10.3389/fped.2024.1489648. eCollection 2024. Front Pediatr. 2024. PMID: 39639950 Free PMC article.
-
State-space modelling using wastewater virus and epidemiological data to estimate reported COVID-19 cases and the potential infection numbers.J R Soc Interface. 2025 Jan;22(222):20240456. doi: 10.1098/rsif.2024.0456. Epub 2025 Jan 8. J R Soc Interface. 2025. PMID: 39772733
References
-
- de Souza A.S., de Freitas Amorim V.M., Guardia G.D.A., Dos Santos F.F., Ulrich H., Galante P.A.F., de Souza R.F., Guzzo C.R. Severe acute respiratory syndrome coronavirus 2 variants of concern: a perspective for emerging more transmissible and vaccine-resistant strains. Viruses. 2022;14 doi: 10.3390/v14040827. - DOI - PMC - PubMed
-
- Hassan A.O., Case J.B., Winkler E.S., Thackray L.B., Kafai N.M., Bailey A.L., McCune B.T., Fox J.M., Chen R.E., Alsoussi W.B., et al. A SARS-CoV-2 infection model in mice demonstrates protection by neutralizing antibodies. Cell. 2020;182:744–753.e4. doi: 10.1016/j.cell.2020.06.011. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

