Development and transdifferentiation into inner hair cells require Tbx2
- PMID: 36687561
- PMCID: PMC9844247
- DOI: 10.1093/nsr/nwac156
Development and transdifferentiation into inner hair cells require Tbx2
Abstract
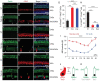
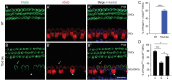
Atoh1 is essential for the development of both outer hair cells (OHCs) and inner hair cells (IHCs) in the mammalian cochlea. Whereas Ikzf2 is necessary for OHC development, the key gene required for IHC development remains unknown. We found that deletion of Tbx2 in neonatal IHCs led to their transdifferentiation into OHCs by repressing 26.7% of IHC genes and inducing 56.3% of OHC genes, including Ikzf2. More importantly, persistent expression of Tbx2 coupled with transient Atoh1 expression effectively reprogrammed non-sensory supporting cells into new IHCs expressing the functional IHC marker vGlut3. The differentiation status of these new IHCs was considerably more advanced than that previously reported. Thus, Tbx2 is essential for IHC development and co-upregulation of Tbx2 with Atoh1 in supporting cells represents a new approach for treating deafness related to IHC degeneration.
Keywords: Ikzf2; Tbx2; cochlea; inner hair cell; outer hair cell; supporting cell.
© The Author(s) 2022. Published by Oxford University Press on behalf of China Science Publishing & Media Ltd.
Figures




Similar articles
-
Revisiting the Potency of Tbx2 Expression in Transforming Outer Hair Cells into Inner Hair Cells at Multiple Ages In Vivo.J Neurosci. 2024 Jun 5;44(23):e1751232024. doi: 10.1523/JNEUROSCI.1751-23.2024. J Neurosci. 2024. PMID: 38688721 Free PMC article.
-
Tbx2 is a master regulator of inner versus outer hair cell differentiation.Nature. 2022 May;605(7909):298-303. doi: 10.1038/s41586-022-04668-3. Epub 2022 May 4. Nature. 2022. PMID: 35508658 Free PMC article.
-
TBX2 specifies and maintains inner hair and supporting cell fate in the Organ of Corti.Nat Commun. 2022 Dec 9;13(1):7628. doi: 10.1038/s41467-022-35214-4. Nat Commun. 2022. PMID: 36494345 Free PMC article.
-
Recent advances in molecular studies on cochlear development and regeneration.Curr Opin Neurobiol. 2023 Aug;81:102745. doi: 10.1016/j.conb.2023.102745. Epub 2023 Jun 23. Curr Opin Neurobiol. 2023. PMID: 37356371 Review.
-
Sensory transduction and frequency selectivity in the basal turn of the guinea-pig cochlea.Philos Trans R Soc Lond B Biol Sci. 1992 Jun 29;336(1278):317-24. doi: 10.1098/rstb.1992.0064. Philos Trans R Soc Lond B Biol Sci. 1992. PMID: 1354370 Review.
Cited by
-
Systematic single cell RNA sequencing analysis reveals unique transcriptional regulatory networks of Atoh1-mediated hair cell conversion in adult mouse cochleae.PLoS One. 2023 Dec 11;18(12):e0284685. doi: 10.1371/journal.pone.0284685. eCollection 2023. PLoS One. 2023. PMID: 38079436 Free PMC article.
-
Mapping the developmental potential of mouse inner ear organoids at single-cell resolution.iScience. 2024 Feb 1;27(3):109069. doi: 10.1016/j.isci.2024.109069. eCollection 2024 Mar 15. iScience. 2024. PMID: 38375227 Free PMC article.
-
Precise genetic control of ATOH1 enhances maturation of regenerated hair cells in the mature mouse utricle.Nat Commun. 2024 Oct 24;15(1):9166. doi: 10.1038/s41467-024-53153-0. Nat Commun. 2024. PMID: 39448563 Free PMC article.
-
Jag1 represses Notch activation in lateral supporting cells and inhibits an outer hair cell fate in the medial cochlea.Development. 2024 Nov 1;151(21):dev202949. doi: 10.1242/dev.202949. Epub 2024 Nov 5. Development. 2024. PMID: 39373109 Free PMC article.
-
Gene therapy restores auditory function and rescues damaged inner hair cells in an aged Vglut3 knockout mouse model.Gene Ther. 2025 Aug 21. doi: 10.1038/s41434-025-00558-1. Online ahead of print. Gene Ther. 2025. PMID: 40841774
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
