Continuous-cropping-tolerant soybean cultivars alleviate continuous cropping obstacles by improving structure and function of rhizosphere microorganisms
- PMID: 36687563
- PMCID: PMC9846356
- DOI: 10.3389/fmicb.2022.1048747
Continuous-cropping-tolerant soybean cultivars alleviate continuous cropping obstacles by improving structure and function of rhizosphere microorganisms
Abstract
Introduction: Soybean continuous cropping will change soil microorganisms and cause continuous cropping obstacles, resulting in a significant yield decline. Different soybean cultivars have different tolerances to continuous cropping, but the relationship between continuous cropping tolerance and soil microorganisms is not clear.
Methods: Two soybean cultivars with different tolerances to continuous cropping were used to study the effects of continuous cropping on soil physical and chemical properties, nitrogen and phosphorus cyclic enzyme activities, rhizosphere soil microbial community and function.
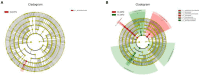
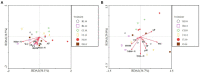
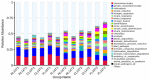
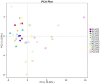
Results: The results showed that the yield reduction rate of a continuous-cropping-tolerant cultivar (L14) was lower than that of a continuous-cropping-sensitive cultivar (L10) under continuous cropping. At R1 and R6 growth stages, soil nutrient content (NH4 +-N, NO3 --N, AP, DOM, TK, and pH), nitrogen cycling enzyme (URE, NAG, LAP) activities, phosphorus cycling enzyme (ALP, NPA, ACP) activities, copy numbers of nitrogen functional genes (AOA, AOB, nirK, nirK) and phosphorus functional genes (phoA, phoB) in L14 were higher than those in L10. Soybean cultivar was an important factor affecting the structure and functional structure of bacterial community under continuous cropping. The relative abundances of Proteobacteria, Bacteroidota, Acidobacteriota and Verrucomicrobiota with L14 were significantly higher than those of L10. The complexity of the soil bacterial community co-occurrence network in L14 was higher than that in L10.
Discussion: The continuous-cropping-tolerant soybean cultivar recruited more beneficial bacteria, changed the structure and function of microbial community, improved soil nitrogen and phosphorus cycling, and reduced the impact of continuous cropping obstacles on grain yield.
Keywords: continuous cropping; enzyme activity; soil bacterial community; soil bacterial function; soil nutrient content; soybean.
Copyright © 2023 Liu, Wang, Yao, He, Sun, Ao, Wang, Zhang, St. Martin, Xie and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Cassava-soybean intercropping alleviates continuous cassava cropping obstacles by improving its rhizosphere microecology.Front Microbiol. 2025 Feb 10;16:1531212. doi: 10.3389/fmicb.2025.1531212. eCollection 2025. Front Microbiol. 2025. PMID: 39996072 Free PMC article.
-
Rhizosphere Soil Bacterial Communities of Continuous Cropping-Tolerant and Sensitive Soybean Genotypes Respond Differently to Long-Term Continuous Cropping in Mollisols.Front Microbiol. 2021 Sep 13;12:729047. doi: 10.3389/fmicb.2021.729047. eCollection 2021. Front Microbiol. 2021. PMID: 34589076 Free PMC article.
-
[Effect of Continuous Cropping on the Physicochemical Properties, Microbial Activity, and Community Characteristics of the Rhizosphere Soil of Codonopsis pilosula].Huan Jing Ke Xue. 2023 Nov 8;44(11):6387-6398. doi: 10.13227/j.hjkx.202211100. Huan Jing Ke Xue. 2023. PMID: 37973120 Chinese.
-
Effects of microbial agent application on the bacterial community in ginger rhizosphere soil under different planting years.Front Microbiol. 2023 Sep 7;14:1203796. doi: 10.3389/fmicb.2023.1203796. eCollection 2023. Front Microbiol. 2023. PMID: 37744902 Free PMC article. Review.
-
Evolutions and Managements of Soil Microbial Community Structure Drove by Continuous Cropping.Front Microbiol. 2022 Feb 28;13:839494. doi: 10.3389/fmicb.2022.839494. eCollection 2022. Front Microbiol. 2022. PMID: 35295291 Free PMC article. Review.
Cited by
-
Comparative analysis of crop rotation systems: the impact of ginger (Zingiber officinale) and sponge gourd (Luffa aegyptiaca) residues on growth of Chinese cabbage (Brassica rapa var. chinensis).Front Plant Sci. 2024 Oct 11;15:1428943. doi: 10.3389/fpls.2024.1428943. eCollection 2024. Front Plant Sci. 2024. PMID: 39464282 Free PMC article.
-
Accumulation patterns of tobacco root allelopathicals across different cropping durations and their correlation with continuous cropping challenges.Front Plant Sci. 2024 Mar 12;15:1326942. doi: 10.3389/fpls.2024.1326942. eCollection 2024. Front Plant Sci. 2024. PMID: 38533406 Free PMC article.
-
Root exudates and rhizosphere microbiota in responding to long-term continuous cropping of tobacco.Sci Rep. 2024 May 17;14(1):11274. doi: 10.1038/s41598-024-61291-0. Sci Rep. 2024. PMID: 38760388 Free PMC article.
-
Microecological Shifts in the Rhizosphere of Perennial Large Trees and Seedlings in Continuous Cropping of Poplar.Microorganisms. 2023 Dec 28;12(1):58. doi: 10.3390/microorganisms12010058. Microorganisms. 2023. PMID: 38257884 Free PMC article.
-
Continuous Cropping of Patchouli Alleviate Soil Properties, Enzyme Activities, and Bacterial Community Structures.Plants (Basel). 2024 Dec 12;13(24):3481. doi: 10.3390/plants13243481. Plants (Basel). 2024. PMID: 39771179 Free PMC article.
References
-
- Amorim H. C. S., Ashworth A. J., Brye K. R., Wienhold B. J., Savin M. C., Owens P. R., et al. . (2020). Soil quality indices as affected by long-term burning, irrigation, tillage, and fertility management. Soil Sci. Soc. Am. J. 85, 379–395. doi: 10.1038/nrmicro283210.1002/saj2.20188 - DOI
-
- Bastian M., Heymann S., Jacomy M. (2009). Gephi: an open source software for exploring and manipulating networks. Int. AAAI Conf. Webl. Soc. Media 3, 361–362. doi: 10.1609/icwsm.v3i1.13937 - DOI
-
- Benitez M. S., Ewing P. M., Osborne S. L., Lehman R. M. (2021). Rhizosphere microbial communities explain positive effects of diverse crop rotations on maize and soybean performance. Soil Biol. Biochem. 159:108309. doi: 10.1016/J.SOILBIO.2021.108309 - DOI
LinkOut - more resources
Full Text Sources
Research Materials

