A culture-, amplification-independent, and rapid method for identification of pathogens and antibiotic resistance profile in bovine mastitis milk
- PMID: 36687564
- PMCID: PMC9852903
- DOI: 10.3389/fmicb.2022.1104701
A culture-, amplification-independent, and rapid method for identification of pathogens and antibiotic resistance profile in bovine mastitis milk
Abstract
Introduction: Rapid and accurate diagnosis of causative pathogens in mastitis would minimize the imprudent use of antibiotics and, therefore, reduce the spread of antimicrobial resistance. Whole genome sequencing offers a unique opportunity to study the microbial community and antimicrobial resistance (AMR) in mastitis. However, the complexity of milk samples and the presence of a high amount of host DNA in milk from infected udders often make this very challenging.
Methods: Here, we tested 24 bovine milk samples (18 mastitis and six non-mastitis) using four different commercial kits (Qiagens' DNeasy® PowerFood® Microbial, Norgens' Milk Bacterial DNA Isolation, and Molzyms' MolYsis™ Plus and Complete5) in combination with filtration, low-speed centrifugation, nuclease, and 10% bile extract of male bovine (Ox bile). Isolated DNA was quantified, checked for the presence/absence of host and pathogen using PCR and sequenced using MinION nanopore sequencing. Bioinformatics analysis was performed for taxonomic classification and antimicrobial resistance gene detection.
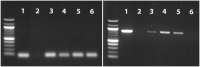
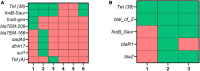
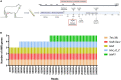
Results: The results showed that kits designed explicitly for bacterial DNA isolation from food and dairy matrices could not deplete/minimize host DNA. Following using MolYsis™ Complete 5 + 10% Ox bile + micrococcal nuclease combination, on average, 17% and 66.5% of reads were classified as bovine and Staphylococcus aureus reads, respectively. This combination also effectively enriched other mastitis pathogens, including Escherichia coli and Streptococcus dysgalactiae. Furthermore, using this approach, we identified important AMR genes such as Tet (A), Tet (38), fosB-Saur, and blaZ. We showed that even 40 min of the MinION run was enough for bacterial identification and detecting the first AMR gene.
Conclusion: We implemented an effective method (sensitivity of 100% and specificity of 92.3%) for host DNA removal and bacterial DNA enrichment (both gram-negative and positive) directly from bovine mastitis milk. To the best of our knowledge, this is the first culture- and amplification-independent study using nanopore-based metagenomic sequencing for real-time detection of the pathogen (within 5 hours) and the AMR profile (within 5-9 hours), in mastitis milk samples. These results provide a promising and potential future on-farm adaptable approach for better clinical management of mastitis.
Keywords: antibiotic resistance; culture-independent sequencing; mastitis; milk; nanopore sequencing technology; rapid diagnosis; staphylococcus aureus; udder infections.
Copyright © 2023 Ahmadi, Khezri, Nørstebø and Ahmad.
Conflict of interest statement
HN was employed by TINE SA. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Assessment of an extraction protocol to detect the major mastitis-causing pathogens in bovine milk.J Dairy Sci. 2011 May;94(5):2171-84. doi: 10.3168/jds.2010-3669. J Dairy Sci. 2011. PMID: 21524507 Clinical Trial.
-
Evaluation of DNA extraction kits for long-read shotgun metagenomics using Oxford Nanopore sequencing for rapid taxonomic and antimicrobial resistance detection.Sci Rep. 2024 Nov 27;14(1):29531. doi: 10.1038/s41598-024-80660-3. Sci Rep. 2024. PMID: 39604411 Free PMC article.
-
16S rRNA nanopore sequencing for rapid diagnosis of causative bacteria in bovine mastitis.Res Vet Sci. 2023 Aug;161:45-49. doi: 10.1016/j.rvsc.2023.06.006. Epub 2023 Jun 10. Res Vet Sci. 2023. PMID: 37321010
-
Antimicrobial resistance of mastitis pathogens.Vet Clin North Am Food Anim Pract. 2012 Jul;28(2):165-85. doi: 10.1016/j.cvfa.2012.03.005. Epub 2012 Apr 28. Vet Clin North Am Food Anim Pract. 2012. PMID: 22664201 Review.
-
Combating Bovine Mastitis in the Dairy Sector in an Era of Antimicrobial Resistance: Ethno-veterinary Medicinal Option as a Viable Alternative Approach.Front Vet Sci. 2022 Apr 4;9:800322. doi: 10.3389/fvets.2022.800322. eCollection 2022. Front Vet Sci. 2022. PMID: 35445101 Free PMC article. Review.
Cited by
-
Bacterial enrichment prior to third-generation metagenomic sequencing improves detection of BRD pathogens and genetic determinants of antimicrobial resistance in feedlot cattle.Front Microbiol. 2024 May 8;15:1386319. doi: 10.3389/fmicb.2024.1386319. eCollection 2024. Front Microbiol. 2024. PMID: 38779502 Free PMC article.
-
Whole genome sequencing in the palm of your hand: how to implement a MinION Galaxy-based workflow in a food safety laboratory for rapid Salmonella spp. serotyping, virulence, and antimicrobial resistance gene identification.Front Microbiol. 2023 Dec 1;14:1254692. doi: 10.3389/fmicb.2023.1254692. eCollection 2023. Front Microbiol. 2023. PMID: 38107857 Free PMC article.
-
Clinical Diagnostics of Bacterial Infections and Their Resistance to Antibiotics-Current State and Whole Genome Sequencing Implementation Perspectives.Antibiotics (Basel). 2023 Apr 19;12(4):781. doi: 10.3390/antibiotics12040781. Antibiotics (Basel). 2023. PMID: 37107143 Free PMC article. Review.
-
Non-Targeted RNA Sequencing: Towards the Development of Universal Clinical Diagnosis Methods for Human and Veterinary Infectious Diseases.Vet Sci. 2024 May 26;11(6):239. doi: 10.3390/vetsci11060239. Vet Sci. 2024. PMID: 38921986 Free PMC article. Review.
-
Short turnaround time of seven to nine hours from sample collection until informed decision for sepsis treatment using nanopore sequencing.Sci Rep. 2024 Mar 19;14(1):6534. doi: 10.1038/s41598-024-55635-z. Sci Rep. 2024. PMID: 38503770 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

