Genomic analysis of Mycobacterium brumae sustains its nonpathogenic and immunogenic phenotype
- PMID: 36687580
- PMCID: PMC9850167
- DOI: 10.3389/fmicb.2022.982679
Genomic analysis of Mycobacterium brumae sustains its nonpathogenic and immunogenic phenotype
Abstract
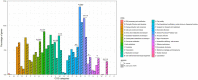
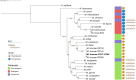
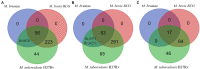
Mycobacterium brumae is a rapid-growing, non-pathogenic Mycobacterium species, originally isolated from environmental and human samples in Barcelona, Spain. Mycobacterium brumae is not pathogenic and it's in vitro phenotype and immunogenic properties have been well characterized. However, the knowledge of its underlying genetic composition is still incomplete. In this study, we first describe the 4 Mb genome of the M. brumae type strain ATCC 51384T assembling PacBio reads, and second, we assess the low intraspecies variability by comparing the type strain with Illumina reads from three additional strains. Mycobacterium brumae genome is composed of a circular chromosome with a high GC content of 69.2% and containing 3,791 CDSs, 97 pseudogenes, one prophage and no CRISPR loci. Mycobacterium brumae has shown no pathogenic potential in in vivo experiments, and our genomic analysis confirms its phylogenetic position with other non-pathogenic and rapid growing mycobacteria. Accordingly, we determined the absence of virulence-related genes, such as ESX-1 locus and most PE/PPE genes, among others. Although the immunogenic potential of M. brumae was proved to be as high as Mycobacterium bovis BCG, the only mycobacteria licensed to treat cancer, the genomic content of M. tuberculosis T cell and B cell antigens in M. brumae genome is considerably lower than those antigens present in M. bovis BCG genome. Overall, this work provides relevant genomic data on one of the species of the mycobacterial genus with high therapeutic potential.
Keywords: diversity; immunogenic; non-pathogenic; nontuberculous mycobacteria; therapeutic.
Copyright © 2023 Renau-Mínguez, Herrero-Abadía, Ruiz-Rodriguez, Sentandreu, Torrents, Chiner-Oms, Torres-Puente, Comas, Julián and Coscolla.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Comparative Genomics and Proteomic Analysis of Four Non-tuberculous Mycobacterium Species and Mycobacterium tuberculosis Complex: Occurrence of Shared Immunogenic Proteins.Front Microbiol. 2016 Jun 7;7:795. doi: 10.3389/fmicb.2016.00795. eCollection 2016. Front Microbiol. 2016. PMID: 27375559 Free PMC article.
-
Nonpathogenic Mycobacterium brumae Inhibits Bladder Cancer Growth In Vitro, Ex Vivo, and In Vivo.Eur Urol Focus. 2016 Apr;2(1):67-76. doi: 10.1016/j.euf.2015.03.003. Epub 2015 Jun 6. Eur Urol Focus. 2016. PMID: 28723453
-
γ Irradiated Mycobacteria Enhance Survival in Bladder Tumor Bearing Mice Although Less Efficaciously than Live Mycobacteria.J Urol. 2016 Jan;195(1):198-205. doi: 10.1016/j.juro.2015.07.011. Epub 2015 Jul 10. J Urol. 2016. PMID: 26165584
-
Expression and regulatory networks of Mycobacterium tuberculosis PE/PPE family antigens.J Cell Physiol. 2019 Jun;234(6):7742-7751. doi: 10.1002/jcp.27608. Epub 2018 Nov 27. J Cell Physiol. 2019. PMID: 30478834 Review.
-
Genetic dissection of immunity to mycobacteria: the human model.Annu Rev Immunol. 2002;20:581-620. doi: 10.1146/annurev.immunol.20.081501.125851. Epub 2001 Oct 4. Annu Rev Immunol. 2002. PMID: 11861613 Review.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

