Acclimation of Nodularia spumigena CCY9414 to inorganic phosphate limitation - Identification of the P-limitation stimulon via RNA-seq
- PMID: 36687591
- PMCID: PMC9846622
- DOI: 10.3389/fmicb.2022.1082763
Acclimation of Nodularia spumigena CCY9414 to inorganic phosphate limitation - Identification of the P-limitation stimulon via RNA-seq
Abstract
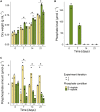
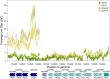
Nodularia spumigena is a toxic, filamentous cyanobacterium capable of fixing atmospheric N2, which is often dominating cyanobacterial bloom events in the Baltic Sea and other brackish water systems worldwide. Increasing phosphate limitation has been considered as one environmental factor promoting cyanobacterial mass developments. In the present study, we analyzed the response of N. spumigena strain CCY9414 toward strong phosphate limitation. Growth of the strain was diminished under P-deplete conditions; however, filaments contained more polyphosphate under P-deplete compared to P-replete conditions. Using RNA-seq, gene expression was compared in N. spumigena CCY9414 after 7 and 14 days in P-deplete and P-replete conditions, respectively. After 7 days, 112 genes were significantly up-regulated in P-deplete filaments, among them was a high proportion of genes encoding proteins related to P-homeostasis such as transport systems for different P species. Many of these genes became also up-regulated after 14 days compared to 7 days in filaments grown under P-replete conditions, which was consistent with the almost complete consumption of dissolved P in these cultures after 14 days. In addition to genes directly related to P starvation, genes encoding proteins for bioactive compound synthesis, gas vesicles formation, or sugar catabolism were stimulated under P-deplete conditions. Collectively, our data describe an experimentally validated P-stimulon in N. spumigena CCY9414 and provide the indication that severe P limitation could indeed support bloom formation by this filamentous strain.
Keywords: alkaline phosphatase; cyanobacterial bloom; diazotroph; polyphosphate; toxin; transcriptomics; transport.
Copyright © 2023 Santoro, Hassenrück, Labrenz and Hagemann.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Insights into the physiology and ecology of the brackish-water-adapted Cyanobacterium Nodularia spumigena CCY9414 based on a genome-transcriptome analysis.PLoS One. 2013;8(3):e60224. doi: 10.1371/journal.pone.0060224. Epub 2013 Mar 28. PLoS One. 2013. PMID: 23555932 Free PMC article.
-
Expression profiling of the bloom-forming cyanobacterium Nodularia CCY9414 under light and oxidative stress conditions.ISME J. 2015 Oct;9(10):2139-52. doi: 10.1038/ismej.2015.16. Epub 2015 Feb 17. ISME J. 2015. PMID: 25689027 Free PMC article.
-
Sedimentation of Nodularia spumigena and distribution of nodularin in the food web during transport of a cyanobacterial bloom from the Baltic Sea to the Kattegat.Harmful Algae. 2019 Jun;86:74-83. doi: 10.1016/j.hal.2019.05.005. Epub 2019 May 30. Harmful Algae. 2019. PMID: 31358279
-
Bacterial diversity and function in the Baltic Sea with an emphasis on cyanobacteria.Ambio. 2007 Apr;36(2-3):180-5. doi: 10.1579/0044-7447(2007)36[180:bdafit]2.0.co;2. Ambio. 2007. PMID: 17520932 Review.
-
Climate change and regulation of hepatotoxin production in Cyanobacteria.FEMS Microbiol Ecol. 2014 Apr;88(1):1-25. doi: 10.1111/1574-6941.12291. Epub 2014 Mar 3. FEMS Microbiol Ecol. 2014. PMID: 24490596 Review.
Cited by
-
Distinctive physiology of polyphosphate-accumulating Beggiatoa suggests an important role in benthic phosphorus cycling.Appl Environ Microbiol. 2025 May 21;91(5):e0233024. doi: 10.1128/aem.02330-24. Epub 2025 Apr 30. Appl Environ Microbiol. 2025. PMID: 40304518 Free PMC article.
References
-
- Bushnell B. (2014). BBMap: a fast, accurate, splice-aware aligner (No. LBNL-7065E). Berkeley, CA: Lawrence Berkeley National Lab.(LBNL).
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

