Mining novel cis-regulatory elements from the emergent host Rhodosporidium toruloides using transcriptomic data
- PMID: 36687612
- PMCID: PMC9853887
- DOI: 10.3389/fmicb.2022.1069443
Mining novel cis-regulatory elements from the emergent host Rhodosporidium toruloides using transcriptomic data
Abstract
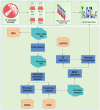
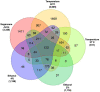
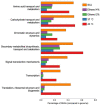
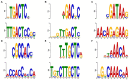
The demand for robust microbial cell factories that produce valuable biomaterials while resisting stresses imposed by current bioprocesses is rapidly growing. Rhodosporidium toruloides is an emerging host that presents desirable features for bioproduction, since it can grow in a wide range of substrates and tolerate a variety of toxic compounds. To explore R. toruloides suitability for application as a cell factory in biorefineries, we sought to understand the transcriptional responses of this yeast when growing under experimental settings that simulated those used in biofuels-related industries. Thus, we performed RNA sequencing of the oleaginous, carotenogenic yeast in different contexts. The first ones were stress-related: two conditions of high temperature (37 and 42°C) and two ethanol concentrations (2 and 4%), while the other used the inexpensive and abundant sugarcane juice as substrate. Differential expression and functional analysis were implemented using transcriptomic data to select differentially expressed genes and enriched pathways from each set-up. A reproducible bioinformatics workflow was developed for mining new regulatory elements. We then predicted, for the first time in this yeast, binding motifs for several transcription factors, including HAC1, ARG80, RPN4, ADR1, and DAL81. Most putative transcription factors uncovered here were involved in stress responses and found in the yeast genome. Our method for motif discovery provides a new realm of possibilities in studying gene regulatory networks, not only for the emerging host R. toruloides, but for other organisms of biotechnological importance.
Keywords: RNA sequencing; biorefineries; cell factory; industrial stress; motifs; sugarcane juice; transcription factor.
Copyright © 2023 Nora, Cassiano, Santana, Guazzaroni, Silva-Rocha and da Silva.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Abdel-Banat B. M. A., Hoshida H., Ano A., Nonklang S., Akada R. (2010). High-temperature fermentation: how can processes for ethanol production at high temperatures become superior to the traditional process using mesophilic yeast? Appl. Microbiol. Biotechnol. 85, 861–867. doi: 10.1007/s00253-009-2248-5, PMID: - DOI - PubMed
-
- Andrews S. (2018). FastQC: a quality control tool for high throughput sequence data, FastQC: a quality control tool for high throughput sequence data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (Accessed October 10, 2022).
-
- Bastian M., Heymann S., Jacomy M. (2009). ‘Gephi: an open source software for exploring and manipulating networks’. In Proceedings of the Third International ICWSM Conference, pp. 361–362.
LinkOut - more resources
Full Text Sources

