Rapid detection of multiple resistance genes to last-resort antibiotics in Enterobacteriaceae pathogens by recombinase polymerase amplification combined with lateral flow dipstick
- PMID: 36687650
- PMCID: PMC9850091
- DOI: 10.3389/fmicb.2022.1062577
Rapid detection of multiple resistance genes to last-resort antibiotics in Enterobacteriaceae pathogens by recombinase polymerase amplification combined with lateral flow dipstick
Abstract
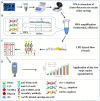
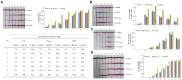
The worrying emergence of multiple resistance genes to last-resort antibiotics in food animals and human populations throughout the food chain and relevant environments has been increasingly reported worldwide. Enterobacteriaceae pathogens are considered the most common reservoirs of such antibiotic resistance genes (ARGs). Thus, a rapid, efficient and accurate detection method to simultaneously screen and monitor such ARGs in Enterobacteriaceae pathogens has become an urgent need. Our study developed a recombinase polymerase amplification (RPA) assay combined with a lateral flow dipstick (LFD) for simultaneously detecting predominant resistance genes to last-resort antibiotics of Enterobacteriaceae pathogens, including mcr-1, blaNDM-1 and tet(X4). It is allowed to complete the entire process, including crude DNA extraction, amplification as well as reading, within 40 min at 37°C, and the detection limit is 101 copies/μl for mcr-1, blaNDM-1 and tet(X4). Sensitivity analysis showed obvious association of color signals with the template concentrations of mcr-1, blaNDM-1 and tet(X4) genes in Enterobacteriaceae pathogens using a test strip reader (R 2 = 0.9881, R 2 = 0.9745, and R 2 = 0.9807, respectively), allowing for quantitative detection using multiplex RPA-LFD assays. Therefore, the RPA-LFD assay can suitably help to detect multiple resistance genes to last-resort antibiotics in foodborne pathogens and has potential applications in the field.
Keywords: Enterobacteriaceae; lateral flow dipstick; rapid detection; recombinase polymerase amplification; resistance genes to last-resort antibiotics.
Copyright © 2023 Lu, Wang, Pan, Gu, Lu, Chen, Zhang, Ye, Xiao, Liu, Tang, Tang, Huang, Fang and Jiang.
Conflict of interest statement
LP was employed by Zhejiang Hongzheng Testing Co., Ltd, Ningbo, Zhejiang, China. XG was employed by Zhejiang Gongzheng Testing Center Co., Ltd, Hangzhou, Zhejiang, China. YT was employed by Hangzhou Tiannie Technology Co., Ltd, Hangzhou, Zhejiang, China. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Adekunle C., Mustapha A., Odewale G., Richard O. (2021). Detection of antibiotic resistance genes among multiple drug resistant Pseudomonas aeruginosa isolated from clinical sources in selected health institutions in Kwara state. Infect. Disord. Drug Target. 21, 53–59. doi: 10.2174/1871526520666201116103625 - DOI - PubMed
-
- Ahmad N., Ali S. M., Khan A. U. (2019). Molecular characterization of novel sequence type of carbapenem-resistant New Delhi metallo-β-lactamase-1-producing Klebsiella pneumoniae in the neonatal intensive care unit of an Indian hospital. Int. J. Antimicrob. Agents 53, 525–529. doi: 10.1016/j.ijantimicag.2018.12.005, PMID: - DOI - PubMed
-
- Anyanwu M. U., Anyogu D. C., Chah K. F., Shoyinka V. S. (2022). Mobile colistin resistance (mcr-1) gene-positive Escherichia coli from chickens in Nigeria is potentially pathogenic and transfers colistin resistance to other organisms. Comp. Clin. Pathol. 31, 323–332. doi: 10.1007/s00580-022-03336-2 - DOI
-
- Ayfan A. K., Macdonald J., Harris P. N., Heney C., Paterson D. L., Trembizki E., et al. (2021). Rapid detection of NDM and VIM carbapenemase encoding genes by recombinase polymerase amplification and lateral flow-based detection. Eur. J. Clin. Microbiol. Infect. Dis. 40, 2447–2453. doi: 10.1007/s10096-021-04267-6, PMID: - DOI - PubMed
LinkOut - more resources
Full Text Sources

