Consumption of a high-fat diet alters transcriptional rhythmicity in liver from pubertal mice
- PMID: 36687679
- PMCID: PMC9845732
- DOI: 10.3389/fnut.2022.1068350
Consumption of a high-fat diet alters transcriptional rhythmicity in liver from pubertal mice
Abstract
Introduction: Childhood obesity is associated with adult obesity, which is a risk factor for chronic diseases. Obesity, as an environmental cue, alters circadian rhythms. The hypothesis of this study was that consumption of a high-fat diet alters metabolic rhythms in pubertal mice.
Methods: Weanling female C57BL/6NHsd mice were fed a standard AIN93G diet or a high-fat diet (HFD) for 3 weeks. Livers were collected from six-week-old mice every 4 h over a period of 48 h for transcriptome analysis.
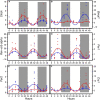
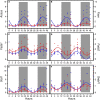
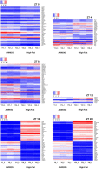
Results and discussion: The HFD altered rhythmicity of differentially rhythmic transcripts in liver. Specifically, the HFD elevated expression of circadian genes Clock, Per1, and Cry1 and genes encoding lipid metabolism Fads1 and Fads2, while decreased expression of circadian genes Bmal1 and Per2 and lipid metabolism genes Acaca, Fasn, and Scd1. Hierarchical clustering analysis of differential expression genes showed that the HFD-mediated metabolic disturbance was most active in the dark phase, ranging from Zeitgeber time 16 to 20. The Kyoto Encyclopedia of Genes and Genomes enrichment analysis of differentially expressed genes showed that the HFD up-regulated signaling pathways related to fatty acid and lipid metabolism, steroid and steroid hormone biosynthesis, amino acid metabolism and protein processing in the endoplasmic reticulum, glutathione metabolism, and ascorbate and aldarate metabolism in the dark phase. Down-regulations included MAPK pathway, lipolysis in adipocytes, Ras and Rap1 pathways, and pathways related to focal adhesion, cell adhesion molecules, and extracellular matrix-receptor interaction. In summary, the HFD altered metabolic rhythms in pubertal mice with the greatest alterations in the dark phase. These alterations may disrupt metabolic homeostasis in puberty and lead to metabolic disorders.
Keywords: circadian rhythms; diet; metabolism; mice; puberty.
Copyright © 2023 Yan, Sundaram, Rust, Palmer, Johnson and Zeng.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
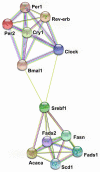
 from curated databases;
from curated databases;  from expirermentally determined;
from expirermentally determined;  from text mining;
from text mining;  from protein homology;
from protein homology;  from co-expression;
from co-expression;  from gene neighborhood;
from gene neighborhood;  from gene fusions. The analysis was performed by using the STRING database (
from gene fusions. The analysis was performed by using the STRING database (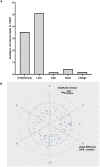


References
-
- Fryar C, Carroll M, Afful J. Division of HEalth and NUtrition Examination Surveys. Prevalence of Overweight, Obesity, and Severe Obesity among Children and Adolescents Aged 2-19 Years: United States, 1963-1965 through 2017-2018. NCHS Health E-Stats. (2020). Available online at: https://www.cdc.gov/nchs/products/index.htm (accessed April 16, 2022).
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

