The cryoEM structure of cytochrome bd from C. glutamicum provides novel insights into structural properties of actinobacterial terminal oxidases
- PMID: 36688035
- PMCID: PMC9846854
- DOI: 10.3389/fchem.2022.1085463
The cryoEM structure of cytochrome bd from C. glutamicum provides novel insights into structural properties of actinobacterial terminal oxidases
Abstract
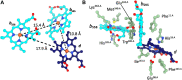
Cytochromes bd are essential for microaerobic respiration of many prokaryotes including a number of human pathogens. These enzymes catalyze the reduction of molecular oxygen to water using quinols as electron donors. Their importance for prokaryotic survival and the absence of eukaryotic homologs make these enzyme ideal targets for antimicrobial drugs. Here, we determined the cryoEM structure of the menaquinol-oxidizing cytochrome bd-type oxygen reductase of the facultative anaerobic Actinobacterium Corynebacterium glutamicum at a resolution of 2.7 Å. The obtained structure adopts the signature pseudosymmetrical heterodimeric architecture of canonical cytochrome bd oxidases formed by the core subunits CydA and CydB. No accessory subunits were identified for this cytochrome bd homolog. The two b-type hemes and the oxygen binding heme d are organized in a triangular geometry with a protein environment around these redox cofactors similar to that of the closely related cytochrome bd from M. tuberculosis. We identified oxygen and a proton conducting channels emerging from the membrane space and the cytoplasm, respectively. Compared to the prototypical enzyme homolog from the E. coli, the most apparent difference is found in the location and size of the proton channel entry site. In canonical cytochrome bd oxidases quinol oxidation occurs at the highly flexible periplasmic Q-loop located in the loop region between TMHs six and seven. An alternative quinol-binding site near heme b 595 was previously identified for cytochrome bd from M. tuberculosis. We discuss the relevance of the two quinol oxidation sites in actinobacterial bd-type oxidases and highlight important differences that may explain functional and electrochemical differences between C. glutamicum and M. tuberculosis. This study expands our current understanding of the structural diversity of actinobacterial and proteobacterial cytochrome bd oxygen reductases and provides deeper insights into the unique structural and functional properties of various cytochrome bd variants from different phylae.
Keywords: CryoEM; electrochemistry; microbiology; oxidases; proton channel.
Copyright © 2023 Grund, Kabashima, Kusumoto, Wu, Welsch, Sakamoto, Michel and Safarian.
Figures




References
LinkOut - more resources
Full Text Sources

