MUC13-miRNA-4647 axis in colorectal cancer: Prospects to identifications of risk factors and clinical outcomes
- PMID: 36688110
- PMCID: PMC9843305
- DOI: 10.3892/ol.2022.13658
MUC13-miRNA-4647 axis in colorectal cancer: Prospects to identifications of risk factors and clinical outcomes
Abstract
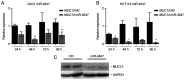
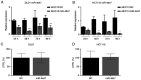
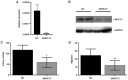
MUC13, a transmembrane mucin glycoprotein, is overexpressed in colorectal cancer (CRC), however, its regulation and functions are not fully understood. It has been shown that MUC13 protects colonic epithelial cells from apoptosis. Therefore, studying MUC13 and MUC13-regulated pathways may reveal promising therapeutic approaches for CRC treatment. Growing evidence suggests that microRNAs (miRs) are involved in the development and progression of CRC. In the present study, the MUC13-miR-4647 axis was addressed in association with survival of patients. miR-4647 is predicted in silico to bind to the MUC13 gene and was analyzed by RT-qPCR in 187 tumors and their adjacent non-malignant mucosa of patients with CRC. The impact of previously mentioned genes on survival and migration abilities of cancer cells was validated in vitro. Significantly upregulated MUC13 (P=0.02) in was observed tumor tissues compared with non-malignant adjacent mucosa, while miR-4647 (P=0.05) showed an opposite trend. Higher expression levels of MUC13 (log-rank P=0.05) were associated with worse patient's survival. The ectopic overexpression of studied miR resulted in decreased migratory abilities and worse survival of cells. Attenuated MUC13 expression levels confirmed the suppression of colony forming of CRC cells. In summary, the present data suggested the essential role of MUC13-miR-4647 in patients' survival, and this axis may serve as a novel therapeutic target. It is anticipated MUC13 may hold significant potential in the screening, diagnosis and treatment of CRC.
Keywords: MUC13; colorectal cancer risk and clinical outcomes; microRNA; translation research.
Copyright: © Sojka et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


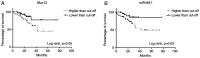




References
-
- Jonckheere N, Skrypek N, Van Seuningen I. Mucins and tumor resistance to chemotherapeutic drugs. Biochim Biophys Acta. 2014;1846:142–151. - PubMed
LinkOut - more resources
Full Text Sources
