In Vivo Genome Editing in Type I and II Methanotrophs Using a CRISPR/Cas9 System
- PMID: 36688528
- PMCID: PMC9942187
- DOI: 10.1021/acssynbio.2c00554
In Vivo Genome Editing in Type I and II Methanotrophs Using a CRISPR/Cas9 System
Abstract
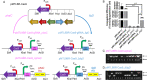
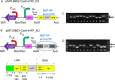
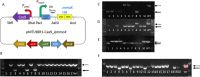
Methanotrophic bacteria are Gram-negative, aerobic organisms that use methane as their sole source of carbon and energy. In this study, we constructed and exemplified a CRISPR/Cas9 genome editing system and used it to successfully make gene deletions and insertions in the type I methanotroph Methylococcus capsulatus Bath and the type II methanotroph Methylocystis parvus OBBP. High frequencies of gene deletions and insertions were achieved in combination with homology-directed repair. In M. parvus OBBP, we also investigated the impact of several parameters on the CRISPR/Cas9 genome editing, where the ligD gene was targeted with various PAM sequences and guide RNA spacer sequences, homology arms of variable length, differences in the duration of mating during conjugation, and exploiting promoters of different strengths to control the expression of cas9 and sgRNA. Although not the first attempt to develop a CRISPR/Cas system in methanotrophs, this work demonstrated for the first time an efficient CRISPR/Cas9 system generating scarless clean gene deletions and insertions in methanotroph genomes.
Keywords: CRISPR; DNA ligase; gene deletion; gene insertion; genome editing; homology-directed repair; methane monooxygenase; methanotrophs; promoter library.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



Similar articles
-
Development of a CRISPR/Cas9 System for Methylococcus capsulatus In Vivo Gene Editing.Appl Environ Microbiol. 2019 May 16;85(11):e00340-19. doi: 10.1128/AEM.00340-19. Print 2019 Jun 1. Appl Environ Microbiol. 2019. PMID: 30926729 Free PMC article.
-
Editing of the Bacillus subtilis Genome by the CRISPR-Cas9 System.Appl Environ Microbiol. 2016 Aug 15;82(17):5421-7. doi: 10.1128/AEM.01453-16. Print 2016 Sep 1. Appl Environ Microbiol. 2016. PMID: 27342565 Free PMC article.
-
An introduction and use of the CRISPR-Cas systems.Prog Mol Biol Transl Sci. 2021;179:1-10. doi: 10.1016/bs.pmbts.2020.12.011. Epub 2021 Jan 27. Prog Mol Biol Transl Sci. 2021. PMID: 33785173
-
Exploring the potential of genome editing CRISPR-Cas9 technology.Gene. 2017 Jan 30;599:1-18. doi: 10.1016/j.gene.2016.11.008. Epub 2016 Nov 9. Gene. 2017. PMID: 27836667 Review.
-
Gene Editing With TALEN and CRISPR/Cas in Rice.Prog Mol Biol Transl Sci. 2017;149:81-98. doi: 10.1016/bs.pmbts.2017.04.006. Epub 2017 May 24. Prog Mol Biol Transl Sci. 2017. PMID: 28712502 Review.
Cited by
-
Enhancing D-lactic acid production from methane through metabolic engineering of Methylomonas sp. DH-1.Microb Cell Fact. 2025 Mar 25;24(1):70. doi: 10.1186/s12934-025-02695-z. Microb Cell Fact. 2025. PMID: 40128822 Free PMC article.
-
Construction of a broad-host-range Anderson promoter series and particulate methane monooxygenase promoter variants expand the methanotroph genetic toolbox.Synth Syst Biotechnol. 2024 Feb 19;9(2):250-258. doi: 10.1016/j.synbio.2024.02.003. eCollection 2024 Jun. Synth Syst Biotechnol. 2024. PMID: 38435708 Free PMC article.
References
-
- Dunfield P. F.The Soil Methane Sink. Greenhouse Gas Sinks; CABI, 2007; p 152.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

