Clinical and Environmental Vibrio cholerae Non-O1, Non-O139 Strains from Australia Have Similar Virulence and Antimicrobial Resistance Gene Profiles
- PMID: 36688638
- PMCID: PMC9927259
- DOI: 10.1128/spectrum.02631-22
Clinical and Environmental Vibrio cholerae Non-O1, Non-O139 Strains from Australia Have Similar Virulence and Antimicrobial Resistance Gene Profiles
Abstract
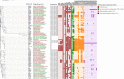
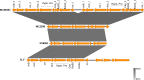
Cholera caused by pathogenic Vibrio cholerae is still considered one of the major health problems in developing countries including those in Asia and Africa. Australia is known to have unique V. cholerae strains in Queensland waterways, resulting in sporadic cholera-like disease being reported in Queensland each year. We conducted virulence and antimicrobial genetic characterization of O1 and non-O1, non-O139 V. cholerae (NOVC) strains (1983 to 2020) from Queensland with clinical significance and compared these to environmental strains that were collected as part of a V. cholerae monitoring project in 2012 of Queensland waterways. In this study, 87 V. cholerae strains were analyzed where O1 (n = 5) and NOVC (n = 54) strains from Queensland and international travel-associated NOVC (n = 2) (61 in total) strains were sequenced, characterized, and compared with seven previously sequenced O1 strains and 18 other publicly available NOVC strains from Australia and overseas to visualize the genetic context among them. Of the 61 strains, three clinical and environmental NOVC serogroup strains had cholera toxin-producing genes, namely, the CTX phage (identified in previous outbreaks) and the complete Vibrio pathogenicity island 1. Phylogenetic analysis based on core genome analysis showed more than 10 distinct clusters and interrelatedness between clinical and environmental V. cholerae strains from Australia. Moreover, 30 (55%) NOVC strains had the cholix toxin gene (chxA) while only 11 (20%) strains had the mshA gene. In addition, 18 (34%) NOVC strains from Australia had the type three secretion system and discrete expression of type six secretion system genes. Interestingly, four NOVC strains from Australia and one NOVC strain from Indonesia had intSXT, a mobile genetic element. Several strains were found to have beta-lactamase (blaCARB-9) and chloramphenicol acetyltransferase (catB9) genes. Our study suggests that Queensland waterways can harbor highly divergent V. cholerae strains and serve as a reservoir for various V. cholerae-associated virulence genes which could be shared among O1 and NOVC V. cholerae strains via mobile genetic elements or horizontal gene transfer. IMPORTANCE Australia has its own V. cholerae strains, both toxigenic and nontoxigenic, that are associated with cholera disease. This study aimed to characterize a collection of clinical and environmental NOVC strains from Australia to understand their virulence and antimicrobial resistance profile and to place strains from Australia in the genetic context of international strains. The findings from this study suggest the toxigenic V. cholerae strains in the Queensland River water system are of public health concern. Therefore, ongoing monitoring and genomic characterization of V. cholerae strains from the Queensland environment are important and would assist public health departments to track the source of cholera infection early and implement prevention strategies for future outbreaks. Understanding the genomics of V. cholerae could also inform the natural ecology and evolution of this bacterium in natural environments.
Keywords: Vibrio cholerae; antimicrobial activity; clinical strains; environmental strains; gene sequencing; virulence factors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Toxigenic Vibrio cholerae strains in South-East Queensland, Australian river waterways.Appl Environ Microbiol. 2023 Oct 31;89(10):e0047223. doi: 10.1128/aem.00472-23. Epub 2023 Oct 6. Appl Environ Microbiol. 2023. PMID: 37800954 Free PMC article.
-
Characterization and Genetic Variation of Vibrio cholerae Isolated from Clinical and Environmental Sources in Thailand.PLoS One. 2017 Jan 19;12(1):e0169324. doi: 10.1371/journal.pone.0169324. eCollection 2017. PLoS One. 2017. PMID: 28103259 Free PMC article.
-
Genomic and Evolutionary Insights into Australian Toxigenic Vibrio cholerae O1 Strains.Microbiol Spectr. 2023 Feb 14;11(1):e0361722. doi: 10.1128/spectrum.03617-22. Epub 2022 Dec 19. Microbiol Spectr. 2023. PMID: 36533913 Free PMC article.
-
Global emergence of environmental non-O1/O139 Vibrio cholerae infections linked with climate change: a neglected research field?Environ Microbiol. 2020 Oct;22(10):4342-4355. doi: 10.1111/1462-2920.15040. Epub 2020 May 15. Environ Microbiol. 2020. PMID: 32337781 Review.
-
Antimicrobial resistance among clinical Vibrio cholerae non-O1/non-O139 isolates: systematic review and meta-analysis.Pathog Glob Health. 2023 May;117(3):235-244. doi: 10.1080/20477724.2022.2114620. Epub 2022 Aug 19. Pathog Glob Health. 2023. PMID: 35983997 Free PMC article.
Cited by
-
Vibrio cholerae O47 associated with a cholera-like diarrheal outbreak concurrent with seasonal cholera in Bangladesh.mSphere. 2025 Apr 29;10(4):e0083124. doi: 10.1128/msphere.00831-24. Epub 2025 Apr 2. mSphere. 2025. PMID: 40172221 Free PMC article.
-
Non-O1/O139 environmental Vibrio cholerae from Northern Cameroon reveals potential intra-/inter-continental transmissions.PLoS Negl Trop Dis. 2025 Apr 3;19(4):e0012890. doi: 10.1371/journal.pntd.0012890. eCollection 2025 Apr. PLoS Negl Trop Dis. 2025. PMID: 40179128 Free PMC article.
-
Genomic insights into clinical non-O1/non-O139 Vibrio cholerae isolates in Japan.Microbiol Spectr. 2025 Aug 5;13(8):e0017525. doi: 10.1128/spectrum.00175-25. Epub 2025 Jun 24. Microbiol Spectr. 2025. PMID: 40552807 Free PMC article.
-
Whole Genome Analysis of a Non-O1, Non-O139 Vibrio cholerae Detected from Human Blood in China.Infect Drug Resist. 2023 Aug 21;16:5453-5461. doi: 10.2147/IDR.S420095. eCollection 2023. Infect Drug Resist. 2023. PMID: 37638066 Free PMC article.
-
The use of next-generation sequencing in personalized medicine.ArXiv [Preprint]. 2024 Mar 6:arXiv:2403.03688v1. ArXiv. 2024. Update in: Methods Mol Biol. 2025;2866:287-315. doi: 10.1007/978-1-0716-4192-7_16. PMID: 38495572 Free PMC article. Updated. Preprint.
References
-
- Camacho A, Bouhenia M, Alyusfi R, Alkohlani A, Naji MAM, de Radiguès X, Abubakar AM, Almoalmi A, Seguin C, Sagrado MJ, Poncin M, McRae M, Musoke M, Rakesh A, Porten K, Haskew C, Atkins KE, Eggo RM, Azman AS, Broekhuijsen M, Saatcioglu MA, Pezzoli L, Quilici M-L, Al-Mesbahy AR, Zagaria N, Luquero FJ. 2018. Cholera epidemic in Yemen, 2016–18: an analysis of surveillance data. Lancet Glob Health 6:e680–e690. doi: 10.1016/S2214-109X(18)30230-4. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

