Development of an Amplicon Nanopore Sequencing Strategy for Detection of Mutations Conferring Intermediate Resistance to Vancomycin in Staphylococcus aureus Strains
- PMID: 36688645
- PMCID: PMC9927139
- DOI: 10.1128/spectrum.02728-22
Development of an Amplicon Nanopore Sequencing Strategy for Detection of Mutations Conferring Intermediate Resistance to Vancomycin in Staphylococcus aureus Strains
Abstract
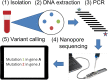
Staphylococcus aureus is a major cause of bacteremia and other hospital-acquired infections. The cell-wall active antibiotic vancomycin is commonly used to treat both methicillin-resistant (MRSA) and sensitive (MSSA) infections. Vancomycin intermediate S. aureus (VISA) variants can arise through de novo mutations. Here, we performed pilot experiments to develop a combined PCR/long-read sequencing-based method for detection of previously known VISA-causing mutations. Primers were designed to generate 10 amplicons covering 16 genes associated with the VISA phenotype. We sequenced amplicon pools as long reads with Oxford Nanopore adapter ligation on Flongle flow cells. We then detected mutations by mapping reads against a parental consensus or known reference sequence and comparing called variants against a database of known VISA mutations from laboratory selection. Each amplicon in the pool was sequenced to high (>1,000×) coverage, and no relationship was found between amplicon length and coverage. We also were able to detect the causative mutation (walK 646C>G) in a VISA mutant derived from the USA300 strain (N384-3 from parental strain N384). Mixing mutant (N384-3) and parental (N384) DNA at various ratios from 0 to 1 mutant suggested a mutation detection threshold of the average minor allele frequency (6.5%) at 95% confidence (two standard errors above mean mutation frequency). The study lays the groundwork for direct S. aureus antibiotic resistance genotype inference using rapid nanopore sequencing from clinical samples. IMPORTANCE Bacteremia mortality is known to increase rapidly with time after infection, making rapid diagnostics and treatment necessary. Successful treatment depends on correct administration of antibiotics based on knowledge of strain antibiotic susceptibility. Staphylococcus aureus is a major causative agent of bacteremia that is also commonly antibiotic resistant. In this work, we develop a method to accelerate detection of a complex, polygenic antibiotic resistance phenotype in S. aureus, vancomycin-intermediate resistance (VISA), through long-read genomic sequencing of amplicons representing genes most commonly mutated in VISA selection. This method both rapidly identifies VISA genotypes and incorporates the most comprehensive database of VISA genetic determinants known to date.
Keywords: Staphylococcus aureus; amplicon sequencing; antibiotic resistance; clinical microbiology; diagnostics; genomics; nanopore sequencing; vancomycin resistance; vancomycin-intermediate Staphylococcus aureus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Staphylococcus aureus response and adaptation to vancomycin.Adv Microb Physiol. 2024;85:201-258. doi: 10.1016/bs.ampbs.2024.04.006. Epub 2024 Jun 1. Adv Microb Physiol. 2024. PMID: 39059821 Review.
-
Insights into the molecular basis of reduced vancomycin susceptibility among three prominent Staphylococcus aureus clonal complexes.Microbiol Spectr. 2024 Aug 6;12(8):e0048624. doi: 10.1128/spectrum.00486-24. Epub 2024 Jun 25. Microbiol Spectr. 2024. PMID: 38916317 Free PMC article.
-
Genetic pathway in acquisition and loss of vancomycin resistance in a methicillin resistant Staphylococcus aureus (MRSA) strain of clonal type USA300.PLoS Pathog. 2012 Feb;8(2):e1002505. doi: 10.1371/journal.ppat.1002505. Epub 2012 Feb 2. PLoS Pathog. 2012. PMID: 22319446 Free PMC article.
-
Impact of vancomycin use trend change due to the availability of alternative antibiotics on the prevalence of Staphylococcus aureus with reduced vancomycin susceptibility: a 14-year retrospective study.Antimicrob Resist Infect Control. 2022 Aug 5;11(1):101. doi: 10.1186/s13756-022-01140-9. Antimicrob Resist Infect Control. 2022. PMID: 35932086 Free PMC article.
-
Clinical implications of vancomycin heteroresistant and intermediately susceptible Staphylococcus aureus.Pharmacotherapy. 2015 Apr;35(4):424-32. doi: 10.1002/phar.1577. Pharmacotherapy. 2015. PMID: 25884530 Review.
Cited by
-
Genomic epidemiology of the primary methicillin-resistant Staphylococcus aureus clones causing invasive infections in Paraguayan children.Microbiol Spectr. 2024 Apr 2;12(4):e0301223. doi: 10.1128/spectrum.03012-23. Epub 2024 Feb 28. Microbiol Spectr. 2024. PMID: 38415665 Free PMC article.
-
Diagnosis and treatment of refractory infectious diseases using nanopore sequencing technology: Three case reports.World J Clin Cases. 2024 Aug 6;12(22):5208-5216. doi: 10.12998/wjcc.v12.i22.5208. World J Clin Cases. 2024. PMID: 39109020 Free PMC article.
-
Epidemiologic analysis of antimicrobial resistance in hospital departments in China from 2022 to 2023.J Health Popul Nutr. 2024 Mar 6;43(1):39. doi: 10.1186/s41043-024-00526-2. J Health Popul Nutr. 2024. PMID: 38449053 Free PMC article.
-
Advances in genotypic antimicrobialresistance testing: a comprehensive review.Sci China Life Sci. 2025 Jan;68(1):130-143. doi: 10.1007/s11427-023-2570-4. Epub 2024 Sep 18. Sci China Life Sci. 2025. PMID: 39300049 Review.
References
-
- Howden BP, Davies JK, Johnson PDR, Stinear TP, Grayson ML. 2010. Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: resistance mechanisms, laboratory detection, and clinical implications. Clin Microbiol Rev 23:99–139. doi:10.1128/CMR.00042-09. - DOI - PMC - PubMed
-
- Chang S, Sievert DM, Hageman JC, Boulton ML, Tenover FC, Downes FP, Shah S, Rudrik JT, Pupp GR, Brown WJ, Cardo D, Fridkin SK, Vancomycin-Resistant Staphylococcus aureus Investigative Team . 2003. Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. N Engl J Med 348:1342–1347. doi:10.1056/NEJMoa025025. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

