A Zinc Finger Motif in the P1 N Terminus, Highly Conserved in a Subset of Potyviruses, Is Associated with the Host Range and Fitness of Telosma Mosaic Virus
- PMID: 36688651
- PMCID: PMC9972955
- DOI: 10.1128/jvi.01444-22
A Zinc Finger Motif in the P1 N Terminus, Highly Conserved in a Subset of Potyviruses, Is Associated with the Host Range and Fitness of Telosma Mosaic Virus
Abstract
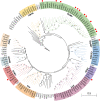
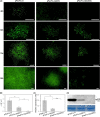
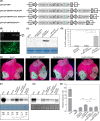
P1 is the first protein translated from the genomes of most viruses in the family Potyviridae, and it contains a C-terminal serine-protease domain that cis-cleaves the junction between P1 and HCPro in most cases. Intriguingly, P1 is the most divergent among all mature viral factors, and its roles during viral infection are still far from understood. In this study, we found that telosma mosaic virus (TelMV, genus Potyvirus) in passion fruit, unlike TelMV isolates present in other hosts, has two stretches at the P1 N terminus, named N1 and N2, with N1 harboring a Zn finger motif. Further analysis revealed that at least 14 different potyviruses, mostly belonging to the bean common mosaic virus subgroup, encode a domain equivalent to N1. Using the newly developed TelMV infectious cDNA clones from passion fruit, we demonstrated that N1, but not N2, is crucial for viral infection in both Nicotiana benthamiana and passion fruit. The regulatory effects of N1 domain on P1 cis cleavage, as well as the accumulation and RNA silencing suppression (RSS) activity of its cognate HCPro, were comprehensively investigated. We found that N1 deletion decreases HCPro abundance at the posttranslational level, likely by impairing P1 cis cleavage, thus reducing HCPro-mediated RSS activity. Remarkably, disruption of the Zn finger motif in N1 did not impair P1 cis cleavage and HCPro accumulation but severely debilitated TelMV fitness. Therefore, our results suggest that the Zn finger motif in P1s plays a critical role in viral infection that is independent of P1 protease activity and self-release, as well as HCPro accumulation and silencing suppression. IMPORTANCE Viruses belonging to the family Potyviridae represent the largest group of plant-infecting RNA viruses, including a variety of agriculturally and economically important viral pathogens. Like all picorna-like viruses, potyvirids employ polyprotein processing as the gene expression strategy. P1, the first protein translated from most potyvirid genomes, is the most variable viral factor and has attracted great scientific interest. Here, we defined a Zn finger motif-encompassing domain (N1) at the N terminus of P1 among diverse potyviruses phylogenetically related to bean common mosaic virus. Using TelMV as a model virus, we demonstrated that the N1 domain is key for viral infection, as it is involved both in regulating the abundance of its cognate HCPro and in an as-yet-undefined key function unrelated to protease processing and RNA silencing suppression. These results advance our knowledge of the hypervariable potyvirid P1s and highlight the importance for infection of a previously unstudied Zn finger domain at the P1 N terminus.
Keywords: HCPro; P1; Potyviridae; Potyvirus; RNA silencing suppression; Zn finger motif; host fitness; telosma mosaic virus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

